Nature's model hobby kit - spliced genes (Part 2)
(This is the 2ndd part of a 3 part series. Click HERE to start reading at Part 1)- It recognizes the AUG start signal in the mRNA and initiates the translation process.
- It traverses the bases/"letters" of the mRNA three "letters" at a time by feeding the codons of the message through the "ticker tape reading machine"
- It recognizes the stop signals UGA, UAA and UAG in the mRNA to know when to terminate the translation process.
- It helps to catalyze the formation of the of the covalent bonds between the growing amino acid chain.
The actual execution of the Genetic Code is not performed by the Ribosome but by a combination of enzymes and a different kind of RNA called tRNA. Remember that the rules of the Genetic Code helps the cell to determine which amino acid to use for each of the 61 possible 3 letter codons.
The CODE which is almost universal in all orders of life, which means that the "programs" encoded in viruses are able to execute in the "human operating system".
Summed up the code looks as follows:
G----G----GGG = Glycine (Gly=G) :Non-polar/Hydrophobic
| GGA = Glycine (Gly=G) :Non-polar/Hydrophobic
| GGC = Glycine (Gly=G) :Non-polar/Hydrophobic
| GGT = Glycine (Gly=G) :Non-polar/Hydrophobic
|
A----GAG = Glutamic acid (Glu=E) :Polar Charged -
| GAA = Glutamic acid (Glu=E) :Polar Charged -
| GAC = Aspartic acid (Asp=D) :Polar Charged -
| GAT = Aspartic acid (Asp=D) :Polar Charged -
|
C----GCG = Alanine (Ala=A) :Non-polar/Hydrophobic
| GCA = Alanine (Ala=A) :Non-polar/Hydrophobic
| GCC = Alanine (Ala=A) :Non-polar/Hydrophobic
| GCT = Alanine (Ala=A) :Non-polar/Hydrophobic
|
T----GTG = Valine (Val=V) :Non-polar/Hydrophobic
GTA = Valine (Val=V) :Non-polar/Hydrophobic
GTC = Valine (Val=V) :Non-polar/Hydrophobic
GTT = Valine (Val=V) :Non-polar/Hydrophobic
A----G----AGG = Arginine (Arg=R) :Polar Charged +
| AGA = Arginine (Arg=R) :Polar Charged +
| AGC = Serine (Ser=S) :Polar Uncharged
| AGT = Serine (Ser=S) :Polar Uncharged
|
A----AAG = Lysine (Lys=K) :Polar Charged +
| AAA = Lysine (Lys=K) :Polar Charged +
| AAC = Asparagine (Asn=N) :Polar Uncharged
| AAT = Asparagine (Asn=N) :Polar Uncharged
|
C----ACG = Threonine (Thr=T) :Polar Uncharged
| ACA = Threonine (Thr=T) :Polar Uncharged
| ACC = Threonine (Thr=T) :Polar Uncharged
| ACT = Threonine (Thr=T) :Polar Uncharged
|
T----ATG = Methionine (Met=M) :Non-polar/Hydrophobic
ATA = Isoleucine (Ile=I) :Non-polar/Hydrophobic
ATC = Isoleucine (Ile=I) :Non-polar/Hydrophobic
ATT = Isoleucine (Ile=I) :Non-polar/Hydrophobic
C----G----CGG = Arginine (Arg=R) :Polar Charged +
| CGA = Arginine (Arg=R) :Polar Charged +
| CGC = Arginine (Arg=R) :Polar Charged +
| CGT = Arginine (Arg=R) :Polar Charged +
|
A----CAG = Glutamine (Gln=Q) :Polar Uncharged
| CAA = Glutamine (Gln=Q) :Polar Uncharged
| CAC = Histidine (His=H) :Polar Charged +
| CAT = Histidine (His=H) :Polar Charged +
|
C----CCG = Proline (Pro=P) :Non-polar/Hydrophobic
| CCA = Proline (Pro=P) :Non-polar/Hydrophobic
| CCC = Proline (Pro=P) :Non-polar/Hydrophobic
| CCT = Proline (Pro=P) :Non-polar/Hydrophobic
|
T----CTG = Leucine (Leu=L) :Non-polar/Hydrophobic
CTA = Leucine (Leu=L) :Non-polar/Hydrophobic
CTC = Leucine (Leu=L) :Non-polar/Hydrophobic
CTT = Leucine (Leu=L) :Non-polar/Hydrophobic
T----G----TGG = Tryptophan (Trp=W) :Polar Uncharged
| TGA = Stop (STOP=*)
| TGC = Cysteine (Cys=C) :Polar Uncharged
| TGT = Cysteine (Cys=C) :Polar Uncharged
|
A----TAG = Stop (STOP=*)
| TAA = Stop (STOP=*)
| TAC = Tyrosine (Tyr=Y) :Polar Uncharged
| TAT = Tyrosine (Tyr=Y) :Polar Uncharged
|
C----TCG = Serine (Ser=S) :Polar Uncharged
| TCA = Serine (Ser=S) :Polar Uncharged
| TCC = Serine (Ser=S) :Polar Uncharged
| TCT = Serine (Ser=S) :Polar Uncharged
|
T----TTG = Leucine (Leu=L) :Non-polar/Hydrophobic
TTA = Leucine (Leu=L) :Non-polar/Hydrophobic
TTC = Phenylalanine (Phe=F) :Non-polar/Hydrophobic
TTT = Phenylalanine (Phe=F) :Non-polar/Hydrophobic
A 3 letter code-word is "read" by following the branches of the tree from the root of the tree down to a leaf node.
For example: For the DNA codon of ATG we would start with the top node of A, then follow the branch of T and end up at the leaf node of Methionine (M).
The execution of the rules prescribed in the Genetic Code is therefore implemented by specialized proteins/enzymes called aminoacyl-tRNA-synthetases of which there are 19 different types encoded (at 69 different locations) in the human genome. They have the task of matching the correct amino acid with the correct transfer RNA (tRNA) adapter molecule of which there are 48 different types encoded (at 500 locations) in the human genome. As these genes a cardinally important in the information processing system of the body, genes that code for the same tRNA (with almost exactly identical sequences) are encoded at various positions like a double redundancy backup system.
As I have already mentioned, a total of almost 500 non-protein coding genes scattered all over the human genome contains the sequence information to produce tRNAs with the exact sequence required the form the secondary structure of the 48 types of tRNAs corresponding to the implemented codon code-words. Instead of possible 61 used throughout nature, the body has cut down on the number of tRNAs required by using a different base to code for T in RNA. The base U (Uracil) (found only in RNA in the place of the letter T found in DNA) is able to complementary bind to more than one kind of base when the mRNA interacts with the tRNA adapter molecule.
Perhaps you now realize why I have colored some of the codons in the above Genetic Code tree in pink and red. Those are the codons for which there are no corresponding tRNA gene in the human genome. The ones in pink, when found in an mRNA message can still work due to the "wobble" base pairing of Uracil, and the ones in red represent the STOP signals for the translation machinery. When the Ribosome encounters one of these, it will come to a standstill due to no matching anti-codon tRNA that carries an amino acid - which will terminate the built up polypeptide and result in the ejection of the protein from the machine.
This lookup of code words reminds me of how the German army used the Enigma code with shared code books to send encrypted messages across insecure radio channels, a story very accurately depicted in movies such as The Imitation Game, Enigma and U-571. Until Alan Turing helped crack the code, the German forces were able to avoid having their communication intercepted.
I urge you to read this Wikipedia article on the Enigma machine...
"During World War II, codebooks were only used each day to set up the rotors, their ring settings and the plugboard....Prior to encryption the message was encoded using the Kurzsignalheft code book. The Kurzsignalheft contained tables to convert sentences into four-letter groups.
 |
| Actual Enigma code book used for an extra level of encryption. |
In the following picture you can see an actual mechanical Enigma machine used for encryption.
I feel this use of a code book is a very good analogy of how the translation of the 4 letter DNA code to 20 letter works. It also uses a "code book" to look up....the Genetic Code Book.
Information is encoded in DNA as complementary bound letters:
A - T (always with 2 hydrogen bonds)
G - C (always with 3 hydrogen bonds)
G - C (always with 3 hydrogen bonds)
A - has a double ring structure
T - has a single ring structure
G - has a double ring structure
T - has a single ring structure
G - has a double ring structure
C - has a single ring structure
This means that there are always 3 rings between the backbone of the DNA ladder when bases are properly paired - a fact that is used by some of the error correcting machinery to locate mismatches:
When A is incorrectly paired with G, there are 4 rings, making the paired DNA bulge.
When C is incorrectly paired with T, there are only 2 rings, making the paired DNA narrower than usual.
When C is incorrectly paired with A, there is indeed 3 rings, BUT, there is a mismatch between C (which normally makes 3 hydrogen bonds) and A (which normally makes 2 hydrogen bonds) - this weakens the bond, allowing it to bend more easily when fed through the MUTS (mismatch repair protein).
The same applies when G is incorrectly paired to T, the 3 bond base G does not bind quite firmly to the 2 bond base T, again allowing the MutS2 dimer to bend the DNA helix in order to detect mismatched bases. In this way the protein machine coded for by the MSH2 gene at position chr2:47630206-47630541 is able to act as a forward error detection and correction code used in communication systems.
(The figure below is simply for illustration and not completely accurate)
In a certain sense the flow of information from the chromosomes to the end product changes hands in the relay race as the gene baton is passed from:
- DNA to pre-RNA (by the DNA directed RNA Polymerase machine)
- pre-mRNA to messenger RNA (mRNA) when the exons are excised and spliced together by the Spliceosome (which contains short RNA sequences used for recognition of splice junctions)
- mRNA to a protein chain of specifically ordered amino acids (by the Ribosome consisting of Ribosomal rRNA used for recognition of the start and stop sites in the message)
- The structure of each of the 20 amino acids are recognized by the 19 different kinds of aminoacyl-tRNA-synthetases and then covalently linked to the 48 tRNA molecules which will carry the amio acids to the Ribosomes.
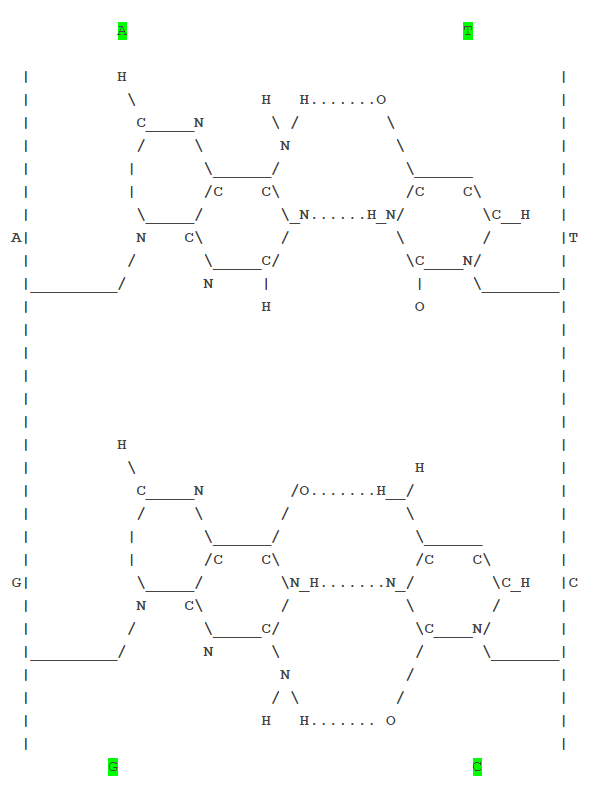
- The 48 tRNA (out of a possible 61 in total) act as adapter molecules, in a similar way that electrical adapter plugs function to convert, say, Australian plugs to South African electrical power outlets. tRNA molecules has the clover leaf shape, where the bottom loop contains the complementary code-word (codon) to what it needs to bind to on the messenger RNA. For example in the case of Methionine which would have the codon of AUG on the messenger RNA strand, its matching tRNA molecule would have UAC in its anti-codon loop in order that they can hybridize / bind to each other as follows:
A - U - G (mRNA)
U - A - C (tRNA)
There are 3 sites on a Ribosome each allowing one codon of 3 bases to be bound (in other word...9 bases are able to bind in parallel).
In this analogy - the Amino acid is the electrical appliance, the tRNA is the adapter plug and the Ribosome binding sites are the power outlets. The aminoacyl-tRNA-synthetase is the user of the appliance which picks the electrical appliance he wants to use and plugs it into the power outlet using the appropriate adapter plug.
You might be wondering why there are only 19 enzymes when there are a total of 20 possible amino acids...
Well, one of these enzyme machines are actually re-purposed to attach 2 different amino acids to the correct tRNAs. This is the enzyme matching Proline and Glutamic Acid.
It links the amino acid Proline to the tRNAs for the codons:
CCG - matching the tRNA anti-codon CGG (in reverse order)
CCA - matching the tRNA anti-codon UGG (in reverse order)
CCC - this is matched by UGG as well due to the base "wobble" of Uracil (U)
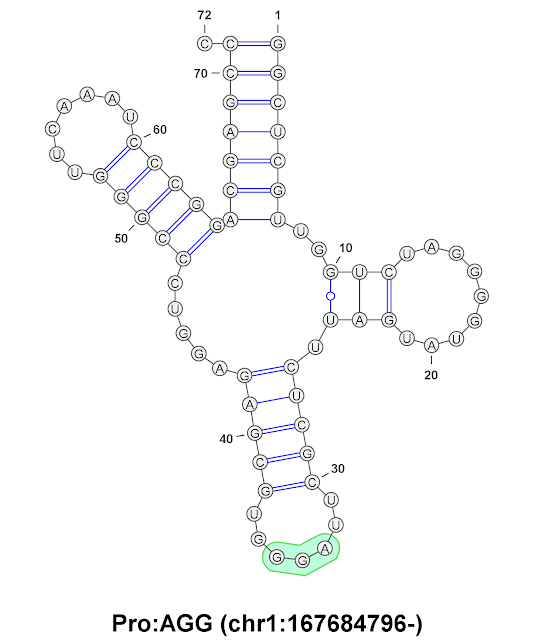
CCU - matching the tRNA anti-codon AGG (in reverse order)
In the following figure is a depiction of the adapter molecule for AGG.
But the same enzyme also knows to differentiate Glutamic Acid and then links it to the tRNAs for the codons:
CUC - matching the tRNA anti-codon GAG (in reverse order)
UUC - matching the tRNA anti-codon GAA (in reverse order)
In the following figure is a depiction of the adapter molecule for CUC.
(Their names are derived from the amino acid letter that they match with)
Alanine (Ala) A
AARS
Homo sapiens alanyl-tRNA synthetase (AARS)
Serine (Ser) S
SARS
Homo sapiens seryl-tRNA synthetase (SARS)
Threonine (Thr) T
TARS
Homo sapiens threonyl-tRNA synthetase (TARS)
Lysine (Lys) K
KARS
Homo sapiens lysyl-tRNA synthetase (KARS)
Arginine (Arg) R
RARS
Homo sapiens arginyl-tRNA synthetase (RARS)
Glutamine (Gln) Q
QARS
Homo sapiens glutaminyl-tRNA synthetase (QARS)
Glutamic acid (Glu) E (GAG/GAA)
Proline (Pro) P (CCG/CCA/CCC/CCT)
EPRS
Homo sapiens glutamyl-prolyl-tRNA synthetase (EPRS)
Aspartic acid (Asp) D
DARS
Homo sapiens aspartyl-tRNA synthetase (DARS)
Asparagine (Asn) N
NARS
Homo sapiens asparaginyl-tRNA synthetase (NARS)
Histidine (His) H
HARS
Homo sapiens histidyl-tRNA synthetase (HARS)
Glycine (Gly) G
GARS
Homo sapiens glycyl-tRNA synthetase (GARS)
Valine (Val) V
VARS
Homo sapiens valyl-tRNA synthetase (VARS)
Isoleucine (Ile) I
IARS
Homo sapiens isoleucyl-tRNA synthetase (IARS)
Leucine (Leu) L
LARS
Homo sapiens leucyl-tRNA synthetase (LARS)
Methionine (Met) M
MARS
Homo sapiens methionyl-tRNA synthetase (MARS)
Phenylalanine (Phe) F
FARSA
Homo sapiens phenylalanyl-tRNA synthetase, alpha subunit (FARSA)
Tyrosine (Tyr) Y
YARS
Homo sapiens tyrosyl-tRNA synthetase (YARS)
Cysteine (Cys) C
CARS
Homo sapiens cysteinyl-tRNA synthetase (CARS)
Tryptophan (Trp) W
WARS
Homo sapiens tryptophanyl-tRNA synthetase (WARS)
These enzymes are the ones to be credited with the "matchmaking" process between the base-4 DNA coding system and the base-20 protein coding system.
I love to learn about codes. Codes are used in modems to transmit information across unreliable and insecure channels. There are error correcting codes, parity codes, data encryption codes and all of them are based on mathematics and in some way or another involve some form of string manipulation. Converting binary data to text basically entails converting base-2 (binary numbers) to base-128 (ASCII) or base-256 (which means there are 255 character representations in that code).
Here is a video I have created of all the tRNA molecules arranges by amino acid and similarity:
(Feel free to pause the video between transitions between different codons for the same amino acid - to notice that while the anti-codon is changing, other sequence parts (particularly the loop on the left) are remaining the same for the same amino acid...explaining how aminoacyl-tRNA-synthetases are able to know when to attach the same amino acid to tRNA's coding with different anti-codons, but which are still associated with the same amino acid)
Einstein said: "God does not play dice." He was right. God plays scrabble.
(Philip Gold)
When the "machine code" needed for a protein contains the 3 letter codon CAG, this codon gets transcribed into the RNA sequence as CAG, and the
tRNA with the anti codon CUG will then base pair with it in the Ribosome 3D printer:
CAG : mRNA (5’ to 3’ direction)
CUG : tRNA (3’ to 5’ direction) These 3 letters are normally in the centre of the anti codon loop of the tRNA.
This means the tRNA will look as follows in the 5’ to 3’ direction :
(Anti codon in 5’ to 3’ direction: GUC)
GGUUCCAUGGUGUAAUGGUAAGCACUCUGGACUCUGAAUCCAGCGAUCCGAGUUCGAGUCUCGGUGGAACCU
A tRNA sequence consists of different parts which contains the reverse complement of other parts of the sequence:
A is the complement of U and
C is the complement of G
The tRNA anti codon CUG always pairs with the mRNA codon CAG in the reverse direction)
1 : GGUUCCA (Reverse compement of 14:UGGAACC)
2 : UG
3 : GUG (Reverse compement of 5:CAC)
4 : UAAUGGUAAG
5 : CAC (Reverse compement of 3:GUG)
6 : U
7 : CUGGA (Reverse compement of 9:UCCAG)
8 : CU CUG AA (Contains the anti codon pairing with the mRNA)
9 : UCCAG (Reverse compement of 7:CUGGA)
10: CGAU
11: CCGAG (Reverse compement of 13:CUCGG)
12: UUCGAGU
13: CUCGG (Reverse compement of 11:CCGAG)
14: UGGAACC (Reverse compement of 1:GGUUCCA)
15: U
This explains why tRNA molecules always forms the distinctive 3-leaf clover shape. It is because there are 3 sequences which are the reverse
complement of each other and will stick together using nature's velcro hydrogen bonds (almost like the rows and columns in a crossword puzzle match to each other).
Searching across the human genome released in 2007, I located 495 tRNA genes across the chromosomes. Here is a table of the number of tRNA genes found in the genome. (This excludes the mitochondrial DNA only found in the circular DNA)
tRNA Gene count by chromosome:
1 106
2 12
3 8
4 3
5 20
6 164
7 23
8 7
9 8
10 2
11 17
12 12
13 5
14 20
15 8
16 31
17 28
18 2
19 10
20 2
21 2
22 1
X 4
This means most tRNA sequences are found on chromosome 1 and 6.
I then arranged the tRNA molecules based on Hamming distance and found that there are about 340
unique versions of these 495 encoded genes. This means that the EXACT tRNA sequence can be found
duplicated EXACTLY on multiple chromosomes.
Just taking the tRNA coding for Aspartic acid for example: (UCSC HG19 Genome browser positions indicated)
Asp:GTC (chr6:27471523+),
Asp:GTC (chr6:27447453+),
Asp:GTC (chr12:125424264-),
Asp:GTC (chr12:125411962-),
Asp:GTC (chr12:96429799+),
Asp:GTC (chr1:161440276-),
Asp:GTC (chr1:161432895-),
Asp:GTC (chr1:161425485-),
Asp:GTC (chr1:161418104-),
Asp:GTC (chr1:161410686-)
The exact same tRNA sequence which contains the anticodon GTC which binds with the Aspartic acid codon GAC on the mRNA strand, is found at least 10 times in the EXACT sequence on 3 different chromosomes 1,6 and 12:
TCCTCGTTAGTATAGTGGTGAGTATCCCCGCCT
GTC (anticodon)
ACGCGGGAGACCGGGGTTCGATTCCCCGACGGGGAG
I have used the program tRNAScan to generate all of the secondary structures of all the tRNA molecules.
This is a movie of all the Transfer RNA molecules which are part of a human.
The chromosome, position and strand direction in the human genome sequence is indicated.
Just have a look at the following 3 tRNA adapter molecules coding for the Amino acid Alanine:
In their anti-codon loops indicated in green) they have the anti-codons:
AGC
CGC
UGC
which will match the 3-base codons from the messenger RNA:
GCT
GCG
GCA
The last tRNA anticodon UGC will also match with
GCG (due to the Uracil in the first position introducing "wobble" by being able to bind with A or G).
The anti-codon is therefore used to discriminate the coding information on the mRNA.
But, there are enzymes called AminoAcyl-tRNA-Synthetases, which match each of the 20 amino acids to their corresponding tRNAs. These enzymes therefore need to find some common identifying sequence on the tRNAs that would indicate that only Alanine needs to be linked to the tRNAs depicted below.





No comments:
Post a Comment
Please leave me a comment