_______________________________________
"Bats Are Natural Reservoirs of SARS-Like Coronaviruses"
Published 28 October 2005
"Severe acute respiratory syndrome (SARS) emerged in 2002 to 2003 in southern China. The origin of its etiological agent, the SARS coronavirus (SARS-CoV), remains elusive. Here we report that species of bats are a natural host of coronaviruses closely related to those responsible for the SARS outbreak."
"Bats are reservoir hosts of several zoonotic (diseases of animal origin) viruses, including the Hendra and Nipah viruses, which have recently emerged in Australia and East Asia, respectively (9–11). Bats may be persistently infected with many viruses but rarely display clinical symptoms (12). These characteristics and the increasing presence of bats and bat products in food and traditional medicine markets in southern China and elsewhere in Asia (13) led us to survey bats in the search for the natural reservoir of SARS-CoV."
"A complete genome sequence was determined directly... The genome organisation of this virus...tentatively named SARS-like corona virus isolate Rp3 (SL-CoV Rp3), was essentially identical to that of SARS-CoV"
This genome sequence was submitted to the international GenBank:
Submitted (23-MAY-2005) Chinese Academy of Science,
Wuhan Institute of Virology, China
It can be found here:https://www.ncbi.nlm.nih.gov/nuccore/DQ071615.1
_______________________________________
"New SARS-like virus can jump directly from bats to humans, no treatment available"
Published 9 Nov 2015
"SARS first jumped from animals to humans in 2002-2003 and caused a worldwide outbreak, resulting in 8,000 cases, including one case in Chapel Hill. With nearly 800 deaths during that outbreak, SARS-CoV presents much like flu symptoms but then can accelerate, compromise breathing and bring on a deadly form of pneumonia. The outbreak was controlled through public health interventions and the original virus was thought to have been extinct since 2004."
"Baric and his team demonstrated that the newly-identified SARS-like virus, labeled SHC014-CoV and found in the Chinese horseshoe bats, can jump between bats and humans by showing that the virus can latch onto and use the same human and bat receptor for entry. The virus also replicates as well as SARS-CoV in primary human lung cells, the preferred target for infection."
“This virus is highly pathogenic and treatments developed against the original SARS virus in 2002 and the ZMapp drugs used to fight Ebola fail to neutralize and control this particular virus,” said Baric. “So building resources, rather than limiting them, to both examine animal populations for new threats and develop therapeutics is key for limiting future outbreaks.”
"Bat SARS-like coronavirus: It’s not SARS 2.0"
New SARS-like virus can 'jump directly from bats to humans without mutating, sparking fears of a future epidemic'
_______________________________________
Published 12 Sept 2018
"This observation highlights the possibility of cross-species transmission of these viruses. These findings strongly suggest the need for continued surveillance of viruses originating from wild animals and promote further research to study the possibility of cross-species transmission of these viruses."
The Corona virus genomes sequences related to this study was submitted 05-JAN-2018 by the Institute of Military Medicine Nanjing, China
and can be found here:
Bat SARS-like coronavirus isolate bat-SL-CoVZXC21, complete genome
Bat SARS-like coronavirus isolate bat-SL-CoVZC45, complete genome
_______________________________________
This article on ABC news was published today 27 January 2020 (19 minutes ago)
Coronavirus is infectious even during incubation and its ability to spread worldwide is strengthening, Chinese authorities say
"The ability of the new corona virus to spread is strengthening and infections could continue to rise, according to the National Health Commission in China, where nearly 3,000 people have been infected and 80 have died."
"The outbreak has prompted widening curbs on movement within China, with Wuhan — the city where the virus is believed to have started — on virtual lockdown, with transports links all but severed except for emergency vehicles."
"The newly identified corona virus has created alarm because there are still many unknowns surrounding it, such as how dangerous it is and how easily it spreads between people. It can cause pneumonia, which has been deadly in some cases."
Now notice how they say..."Newly identified", although scientists from exactly that same city Wuhan in China had already published their findings on 28 October 2005 indicating that a corona virus hosted in bats has the potential of creating a worldwide pandemic when (not if) it jumps across species.
The full genome sequence of this Wuhan Corona virus has now been published by Chinese scientists from Shanghai Public Health Clinical Center & School of Public Health, Fudan University, Shanghai, China on 5 January 2020:
Wuhan seafood market pneumonia virus isolate Wuhan-Hu-1, complete genome
it is referred to in a paper:
"A novel coronavirus associated with a respiratory disease in Wuhan of Hubei province, China"
To read more about this sequence and its publication:
Novel coronavirus complete genome from the Wuhan outbreak now available in GenBank
where they show it is related to the original SARS and the recent MERS (Middle East Respiratory Syndrome)
_______________________________________
In order to learn more about Corona viruses please refer to the following excellent articles:
"Scientists in China sequenced the virus’s genome and made it available on Jan. 10, just a month after the Dec. 8 report of the first case of pneumonia from an unknown virus in Wuhan. In contrast, after the SARS outbreak began in late 2002, it took scientists much longer to sequence that coronavirus. It peaked in February 2003 — and the complete genome of 29,727 nucleotides wasn’t sequenced until that April."
"The progenitor virus itself was almost certainly one that circulates harmlessly in bats (as SARS does) but has an “intermediate reservoir” in one or more animals that come into contact with people, Andersen said. Presumably, that reservoir is one of the species of animals at the Wuhan market thought to be ground zero for the outbreak. The ancestor of 2019-nCoV existed in that species for some unknown time, never infecting people, until by chance a single virus acquired a mutation that made it capable of jumping into and infecting humans."
"Unfortunately, genetic analysis can’t identify what animal species the coronavirus jumped from into humans. But an analysis by a team from the Wuhan Institute of Virology, posted to the preprint server bioRxiv, determined that the genome of this coronavirus (the seventh known to infect humans) is 96% identical to that of a bat coronavirus, suggesting that species is the original source."
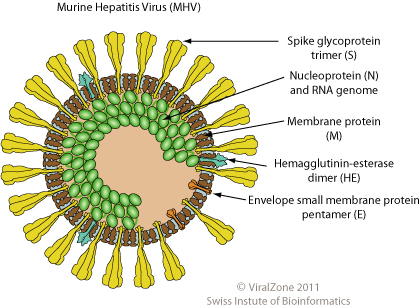
Here is an example of how the coronavirus (which gets its name from the Spike proteins on its surface making it look like a corona (of the sun) or a crown.
_______________________________________
Now, lets do some DNA sequence comparisons between the different published sequences of the
Human SARS Coronavirus (from the 2002-2004 outbreak)
Published on 31 Aug 2004
29751 bases long
BAT related Corona virus (SL-CoV Rp3) (from Horseshoe bat)
Published in 23 May 2005
29736 bases long
Rhinolophus affinis coronavirus isolate LYRa11 (from Horseshoe bat)
Published in 22 Aug 2013
29805 bases long
Bat SARS-like coronavirus isolate bat-SL-CoVZXC21
Published 5 Jan 2018
29732 bases long
Wuhan seafood market pneumonia virus isolate 1 Wuhan-Hu-1 (From the current outbreak)
Published 5 Jan 2020
29903 bases long
Comparing each of the virus proteins will result in a different level of similarity depending on how much the genome had mutated over the years in that part of the RNA genome. The Coronavirus is a single stranded RNA (positive strand) virus of between 29000 and 30000 RNA bases in length. Due to being a single stranded RNA virus its genome is much more prone to RNA letter changes than a double-stranded DNA virus (such as Measles), which has a backup strand in case of mutations.
The Coronavirus life cycle all takes place in the cytoplasm of the host organism's cells and does not need to enter the cell nucleus at all. It carries its own RNA dependent RNA polymerase (copy machinery) and is therefore able to make due without the host's DNA replication machinery. When I compare this copy machine between the different corona virus strains I get the following result:
Using the Dayhoff78 PAM (Percent Accepted Mutation) matrix and calculating the Local Edit distance between the corresponding ORF1ab polyprotein which contains the polymerase protein, I obtained the following percent similarities:
Human SARS Coronavirus to Wuhan-Hu-1
Published on 31 Aug 2004
91.6 % similar
BAT related Corona virus (SL-CoV Rp3) to Wuhan-Hu-1
Published in 23 May 2005
87.7 % similar
Rhinolophus affinis coronavirus isolate LYRa11 (from Horseshoe bat) to Wuhan-Hu-1
Published in 22 Aug 2013
87.9 % similar
Bat SARS-like coronavirus isolate bat-SL-CoVZXC21 to Wuhan-Hu-1
Published 5 Jan 2018
97.4 % similar
Clearly the 2018, CoVZXC21 strain of the virus is very similar to the current Wuhan strain.
And when you compare the Envelope (E) protein you actually get 100% similarity.
And when you compare the Membrane (M) protein you get 99.3% similarity.
When I do a different kind of comparison, by overlaying the genomes of the original 2004 SARS virus on top of the new Wuhan virus (but shifting the SARS virus genome by 69 bases to the left with respect to the start position of the Wuhan virus), and I highlight the differences in Magenta colour, one can get an idea of the amount of mutations that happened over the pas 16 years in this RNA genome.
This is shown at a width of 101 bases.
Zooming in to 47 bases width:
When you create a phylogenetic tree based on the Nucleocapsid, Membrane and Envelope proteins, one can see how different corona viruses of Bat origin are related:
And based on the Spike protein, which binds to the cell receptor in the host cell in order to unlock the cell-door it is most closely related to the 2018 bat strain.
Looking at the structural genes in the genome at a width of 69 bases per line:
_______________________________________
When you zoom out to 161 bases width and colour the codons in the first non-structural gene according to the amino acids coded for by the different codons in the Wuhan genome, the genome looks as follows:
When you zoom out to 161 bases width and colour the codons in the first 23000 or so bases according to the hydrophobic properties of the amino acids coded for by the different codons in the Wuhan genome, the genome looks as follows:
When you look at the protein polypeptide sequence after being translated into amino acids: (for the ORF1ab poly-protein)
7096 amino acids long translated from an open reading frame (start-stop bases) of 21291 RNA genome bases,
The protocol on how to test for Wuhan virus via real time Polymerase Chain Reaction is outlined here.
For a complete comparison between the 2003 SARS virus and the 2020 SARS-COV-2 Virus
look at this link.