Nature's model hobby kit - spliced genes (Part 3)
(This is the 3rd part of a 3 part series. Click HERE to start reading at Part 1)I am drifting off-topic a bit for a reason. I wanted to give you a background of how these encoded "program" instructions we call genes are used in order manufacture different kinds of working protein machines that fall into a few broad classes:
enzymes (such as the digestive Pepsin in the stomach),
hormones (such as Insulin in the blood),
transporter proteins (such as hemoglobin that carry oxygen),
antibodies (of the immune system),
structural building blocks (such as collagen, keratin and elastin),
motor proteins which convert chemical energy into movement (such as actin and myosin in muscles),
receptors (such as the light detecting Rhod-opsin in the retina),
signalling proteins (such as the G-proteins that transmit signals into cells) and
storage proteins (such as albumin and casein which stores energy reserves)
Then there are also a wide variety of protein machines involved in the DNA copy and protein manufacturing process which are indispensable and are encoded from the same DNA that they help to copy and read. These are mostly enzymes which work like machines to catalyze the reactions. If just the enzymes are listed which take part in DNA reproduction (i.e. replication) and repair the list already tops 72 proteins.
Which brings me to the point I wanted to explain in the first place...how are these "machines" assembled? As opposed to bacterial genes and some viral genes which encodes multiple proteins on the same gene, a concept known as polycistronic, eukaryotic multi-cellular organisms have their "recipes" encoded as split-up "modules" called exons and encode only a single protein or regulatory RNA per gene - a concept known as monocistronic.
You can think of a gene as a file on a computer hard drive and the chromosomes as folders. In the following picture you can see a graphical representation of the folders and files on my Surface Tablet.

A disk operating system always tries to find a contiguous region of disk drive where it can store the binary data of the file, but it cannot always find a large enough unused region and have to split the file data up into multiple parts which it writes in-between the data for other files. The disk therefore becomes fragmented and you have to run the program Defrag every once-and-a-while to optimize the disk access. Computer files on a disk takes longer to access when the separate parts of the files have to be fetched all over the place. It is therefore advantageous that the files be located as close together as possible.
 |
| A picture of a very fragmented hard drive. File access will be slow. |
In the following picture is a 2D representation of a part of Chromosome 2. Each of the colorored areas represent the transcribed regions of a number of genes. Some genes are large and others are small. The gene which encodes for human Collagen is labeled as COL5A2 in the picture below. The information of the genes on the DNA can be encoded on thew positive strand and then be read in the forward direction, OR, they can also be encoded on the negative strand and then be read in the opposite direction. A specific gene cannot be read in both directions. Only one of the 2 directions produce a working protein or regulatory RNA. It does sometimes happen that genes which work together in the same biochemical pathway are encoded back to back on the DNA: one running in the positive and another running in the negative direction, BUT where they both share the same promoter region and can be switched on by the same transcription factor protein. Many genes work this way. You can think of it as working like a central control switch in a power plant, activating multiple processes by the same input.
The DNA which must be "read" (by the RNA Polymerase II protein machine) in order to produce the primary RNA transcript or pre-RNA is 147,965 bases in length. Even though not all of these bases are actually going to be used, all of the bases have to be copied into RNA and then further processed before being exported through the pores of the nucleus.
Having a shorter and more compact gene to transcribe would certainly speed things up, but slowing things down may also be part of the way the body regulates how much collagen is actually produced.
Just like a movie editor then cuts and splices a film, the Spliceosome machine in the nucleus then performs editing on the pre-RNA by removing the introns from the transcript to produce mature messenger RNA containing only the exons.
If we now only show the exons of the same genes on Chromosome 2, the same picture would look as follows: (the transcript region is highlighted in a red frame)
You can now see the short scattered exons (depicted in a cyan color) which are spliced together to form the mRNA which will be used as the recipe to "print" the protein.
Only 6930 bases of the originally transcribed 147965 bases are actually used in the mRNA.
This is only 4.7% of the original DNA which was copied. This is like buying 148 meters of electrical wiring when you only want to use 7 meters of it.
We still do not know fully what all the removed parts are used for. In some instances more than one gene are actually transcribed as part of the same transcript where one gene's exons forms part of another gene's intronic regions. Regulatory RNA's can also form part of the intronic mRNA.
One thing is very important...the reading frame (in other words, the sequence of 3 base codons by which the protein is encoded) may not be broken by splicing errors. The reading frame must be maintained. If the splicing machinery, slips by even one base, it will result in all of the downstream codons to be misread as coding for different amino acids, and cause a frame shift mutation. This could result in a truncated protein (due to the introduction of a stop codon) or a protein which fails to fold properly and not functioning correctly as the required enzyme.
I like to think of a spliced gene as the molded plastic frame which contains the parts of a model airplace. The parts of the plane can be quickly mass produced as plastic molded frames which acts as scaffolding holding the "exons" in place which will be used to build the body of the model airplane (representing the enzyme). Again, the parts which are kept are the "exons" and the parts which are left behind are the "introns".
Since the exons represent the codons which encode the protein information, the code/markings indicating where introns start and end are encoded as part of the intron bases and follows a specific coding scheme:
The start of an intron is marked by a consensus sequence: GUAAG or GU in most cases. GTAAG when using DNA letters.
The end of the intron is marked by a consensus sequence AG.
This means the start of the part which is "not used" starts with GT and ends with AG.
It is depicted in the picture below:
EXON1|GT........intron.........A........AG|EXON2|GT........intron.........A........AG|EXON3
One could think of each level of decoding or unpacking of the information as a layer. There are clearly defined sequences which mark the beginning and end of each layer of coding information. Each layer's start and termination markers are different based on the protein machine responsible for decoding it.
If we took a few steps back and look at the entire genome, we will notice the following:
- The 3.2 billion letters of information in the human genome is divided into 23 distinct chromosomes (like folders/directories on a disk) of which there is a backup copy resulting in 46 chromosomes in total (making it 3.2 x 2 = 6.4 billion). (In truth the X and Y chromosome in males do not have backup copies since a male has only 1 X and 1 Y chromosome)
- When the DNA is replicated before cell multiplication (i.e. the disk is duplicated), there are special sequences called replicator sequences at the origins of replication which binds the proteins which "unzips" the double strands to allow the DNA copy machinery to accurately back up the information into sister chromosomes during cell division. There are about 30000 such sequences scattered across the chromosomes.
- Each chromosome contains many genes scattered like "files" across each chromosome. Chromosome 1 is about 249 Megabases in length while Chromosome 22 is about 51 Megabases in length.
- Each gene or "file" is stored as regions on the chromosomes which are either continuously transcribed or on demand. Parts of the "files" exist as smaller fragments called exons which are spliced together to form the RNA message read from the DNA.
- A promoter sequence as well as activator sequence indicates where the RNA Polymerase must start to transcribe the gene into RNA.
The end of the gene is marked by a special DNA sequence which forms RNA which can base-pair with itself to form an RNA secondary structure representing a brake or stop-signal telling it when the gene is fully transcribed and to release the transcript. Some promoter sequences are known as the TATA box as this is where the Tata-binding protein binds which recruits the copy machine to the start site.
The 3' (3 Prime) untranslated region (near the termination of the gene) encodes for elaborate 2D RNA Secondary structures assisting in braking the RNA Polymerase.
Here is the structure for the Collagen gene's 3' untranslated region.
Here is the structure for the Collagen gene's 3' untranslated region.
If you are wondering what the term 5-prime (5') and 3-prime (3') means: It refers to the number of the Carbon atoms in the ribose or deoxyribose sugar molecules. Ribose is a pentose sugar because it consists of 5 carbons in a ring.
The carbon atoms are numbered clockwise starting with the one on the right. As you can see in the picture, the difference between DNA (Deoxyribonucleic acid) and RNA (Ribonucleic acid) is basically at the 2' (2 prime) carbon where an Oxygen has been removed from the OH, leaving only an H. This means DNA has been "de-Oxygenized" at the 2' carbon.Furthermore, DNA is always only extended in the 5' to 3' direction when it is synthesized by the DNA polymerase machines. This means a Phosphate connected to the 5' Carbon side of a subsequent nucleotide binds to the hydroxyl group at the 3' Carbon position.
A nucleotide is the unit consisting of the Phosphate-sugar backbone and the base.
When the RNA Polymerase transcribes the DNA into RNA, it extends the RNA message by covalently binding the Phosphates (PO4) on the 5' carbon side of a new nucleotide to the Hydroxyl (OH) on the 3' carbon side.
Each of the "machines" on the production line is only concerned with its role in the information processing pipeline.
- The DNA directed RNA Polymerase which produces the primary transcript is only concerned with accurately copying the gene letters from the transcription start site marked by the promoter up to the terminator. It does not even know (or need to know) about the exons encoded within the transcript or the special sequences marking the introns, and it is oblivious to any underlying codon structure encoding proteins.
- The Spliceosome is only concerned with identifying where each intron starts and ends in order to accurately remove the introns and join together the exons. It often happens that bases which are part of the same amino acid's codon lie on different exons.
eg. The first base of a 3 letter codon might be the last letter of the first exon while the last 2 bases of the same codon lie thousands of bases away on the start 2 bases of the second exon. - The Ribosome translating the 3 letter codons in the mRNA into protein is totally oblivious to the fact that the codons it is reading were once part of many separate exons spliced together.
In the following picture I have marked the positions of the amino acids which were coded from a codon at the split point of 2 exons in red. Hydrophobic amino acids are in pink, polar in blue, negatively charged polar in green and positively charged polar in cyan. The row lengths are here 35 amino acids wide.
 |
| Protein primary sequence of Collagen gene |
As soon as the number of amino acids in a row is made equal to 36, the amino acids originating from the exons divisions suddenly line up. Indicating that a lot of the exon lengths are multiples of 36 x 3 = 108 bases in length.
 |
| Protein primary sequence of Collagen spaced at 36 amino acids per row |
There are a few nucleoproteins which consist of protein bound with RNA. The sequence of the RNA act as a recognition template for specific sequences on the DNA. This allows the protein to home-in like a heat seeking missile to the target site where it needs to catalyse a reaction.
A few uses of RNA for binding to/synthesizing specific sequences in DNA comes to mind:
- The Ribosome consists of rRNA and proteins allowing it to recognize specific sequences where Translation needs to start and end. (This is especially true for bacteria which recognizes the Shine-Dalgarno sequence AGGAGG)
- The Spliceosome consists of RNA and proteins allowing it to recognize the start, end and branch point of an intron to be removed (in specific the sequences GT and AG)
- Telomerase enzyme synthesizes the repeated sequence TTAGGG at the end of chromosomes (the telomeres) by using RNA as a template
- Transfer/t-RNA recognizes the specific codons using the anti-codon loop of RNA
- Short "interferon" RNAs complementary binds to mRNA target molecules in order to inhibit the production of protein in a negative feedback loop of regulation.
Here is a picture of how the Spliceosome recognizes the start and end of introns using short nuclear RNA bound to proteins and are collectively known as small nuclear ribonucleic proteins (snurps).
Remember how I compared the machinery which recognizes the splice sites (GT/AG) with the clapboard used in movie making to synchronize the audio and video tracks:
In film one needs to accurately synchronize the audio and video streams and a trusted piece of slate clap board used to be employed to accomplish this task. The clapping sound would provide the audio marker when the clap board is closed on film. This simple means of synchronization, together with the date, the scene ("gene") and the take would provide the film editor with enough information to allow him to accurately join or "splice" the pieces of film together whilst also adding the audio track to the film."
In film one needs to accurately synchronize the audio and video streams and a trusted piece of slate clap board used to be employed to accomplish this task. The clapping sound would provide the audio marker when the clap board is closed on film. This simple means of synchronization, together with the date, the scene ("gene") and the take would provide the film editor with enough information to allow him to accurately join or "splice" the pieces of film together whilst also adding the audio track to the film."
 |
I am now going to explain the splicing process using an example of a well known gene product : Opsin
In order for the eye to be able to distinguish between different colours, an eye needs to be able to trigger electrical signals in different neurons leading to the brain depending on the colour being observed.
How is this possible ?
I do not want to go into too much detail.
There is an excellent article that you can read on the subject of colour perception : www.webexhibits.org.
There is an excellent article that you can read on the subject of colour perception : www.webexhibits.org.
Also have a look at the following Wikipedia article on Opsins.
If you want to find out how different colour blindness affects how an image is perceived, you can have a look at this colour blindness simulator.
- Different colours are essentially different wavelengths of light waves hitting the retina of the eye
- The eye has four light detector molecules:
Red / Long wavelength light sensitive opsin (in the cone cells of the eye)
Green / Medium wavelength light sensitive opsin (in the cone cells of the eye)
Blue / Short wavelength light sensitive opsin (in the cone cells of the eye)
Night vision / "Wavelength insensitive" rhodopsin (in the rod cells of the eye) - Each light detecting receptor contains a Vitamin A (Retinol) based molecule called a Chromophore which basically changes shape when light waves of a specific frequency/wavelength falls on it. It's shape remains altered just long enough to set off a domino effect which eventually triggers a voltage pulse in a neuron/nerve cell leading to the retinal ganglion cells which does contrast processing before passing the signal on to the visual cortex of the brain.
- Your brain "sees" colour by mixing the signals it is getting from the different colour detectors into the perceived colour of the scene in front of it
The following picture of the Vitamin-A based chromophore molecule shows how the molecule changes shape in response to a light photon of a specific energy (determined by its wavelength).
The Long Wavelength Sensitive (LWS) "Red detector" only reacts to light of long wavelengths with low energy in its photons
It is coded for by the OPN1LW Opsin gene on Chromosome X at position chrX:153409725-153424507
Its maximum sensitivity is at 560 nm wavelength, in the yellow-green region of the electromagnetic spectrum.
The Medium Wavelength Sensitive (MWS) "Green detector" protein only reacts to light of medium wavelengths
It is coded for by the OPN1MW Opsin gene on Chromosome X at position chrX:153448085-153462352
Its maximum sensitivity is at 530 nm, in the green region of the electromagnetic spectrum.
and
the Short Wavelength Sensitive (SWS) "Blue detector" protein only reacts to light of short wavelengths with high energy in its photons
It is coded for by the OPN1SW Opsin gene on Chromosome 7 at
position chr7:128412543-128415844
Its maximum sensitivity is at 430 nm, in the blue region of the electromagnetic spectrum.
The Chromophore acts like a mechanical actuator which detects single photons of light. When a photon falls on the Chromophore it becomes straightened very fast almost like a "Jack in the box". And it transfers its mechanical energy to G-receptor proteins in a cascade. What is particularly interesting about the Opsin protein's use of the chromophore is the fact that it is NOT different Chromophore molecules that determines at what frequency it reacts and becomes straightened - it is the "stiffness" of the protein amino acid chain holding the Chromophore in its place. The chromophore is only covalently bonded on its one end to the protein, while the other end is held in place by hydrogen bond attraction.
The Blue Opsin molecule seems to be more "stiff" and the Chromophore needs to receive light with higher energy (shorter wavelength) in order for it to be deformed.
The Red Opsin molecule seems to be less "stiff" (or has more damping at high frequencies) and the Chromophore needs to receive light with lower energy (longer wavelength) in order for it to be deformed. Higher energy ("blue") photons does not excite this receptor. I imagine that a mechanism similar to a shock-absorber on a car must be at work. If you slowly depress a shock absorber, you are actually able to depress it, but as soon as you quickly (and with much force) try to depress it, the shock absorber does not want to budge.
The specific SHAPE of the Opsin protein therefore determines the specific frequency to which the Opsin will react. There is a fine tuning going on and the shape that the protein will fold into, is determined by the specific DNA letter sequence coding for the correct amino acids which will for part of the protein chain.
In the following picture you can see a representation of the "Red" Long Wave sensitive light receptor protein. The Purple molecule anchored in the center of the Opsin is the Chromophore responsible for reacting to incoming photons.
 |
| Protein Databank Id: 1KPX |
This ENTIRE shape is determined by the specific hydrophobic/polar nature of the amino acids in the protein chain.
When the amino acids are coloured according to their affinity to co-exist with water (hydrophobicity), the same molecule looks as follows:
 |
| "Cartoon" representation of the LWS Opsin receptor with Chromophore in purple |
Because the Opsin receptors are embedded in the oily membrane of the retina, the hydrophobic sides of the protein needs to be on the outside in contact with the lipid (oily) membrane. That is why the pink hydrophobic amino acids are on the outside. When the Chromophore deforms it produces a "mechanical pulse" carrying the signal through the cell membrane of the Retina cells.
Because molecules consist of atoms with spherical electron clouds around them, the "real life" "Red sensitive" Opsin molecule actually looks more like this:
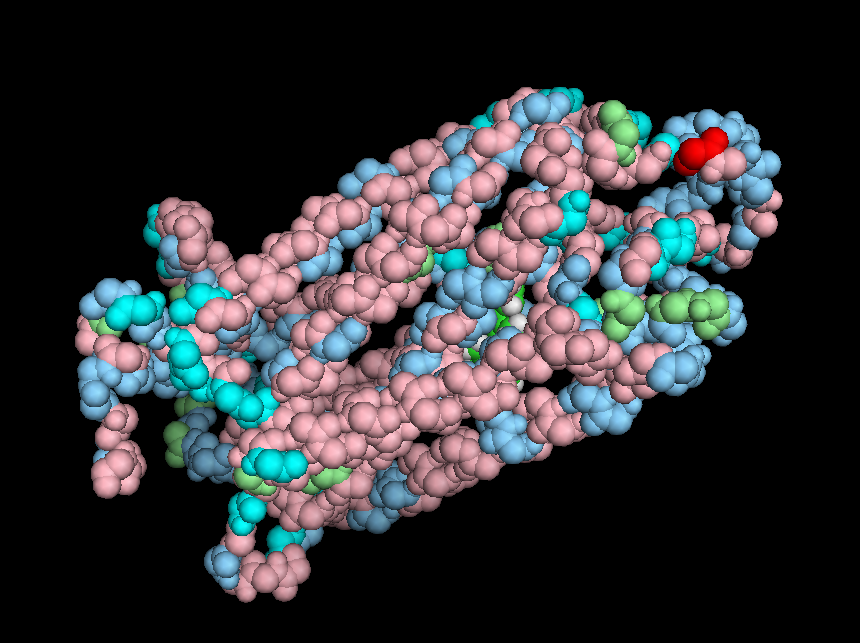 |
| Main amino acid chain space fill representation of the LWS Opsin receptor |
Now that you have a background on the specific protein being made by the Ribosome machinery, lets now work our way backwards through the "Central Dogma" of biology towards how this information is encoded in the Human genome.
The primary sequence of amino acids which folds into the tertiary protein structure seen above, looks as follows when depicted in 2D table format:
Here is a sequence representation of the Opsin protein:
MAQQWSLQRLAGRHPQDSYEDSTQSSIFTYTNSNSTRGPFEGPNYHIAPR
WVYHLTSVWMIFVVTASVFTNGLVLAATMKFKKLRHPLNWILVNLAVADL
AETVIASTISIVNQVSGYFVLGHPMCVLEGYTVSLCGITGLWSLAIISWE
RWLVVCKPFGNVRFDAKLAIVGIAFSWIWSAVWTAPPIFGWSRYWPHGLK
TSCGPDVFSGSSYPGVQSYMIVLMVTCCIIPLAIIMLCYLQVWLAIRAVA
KQQKESESTQKAEKEVTRMVVVMIFAYCVCWGPYTFFACFAAANPGYAFH
PLMAALPAYFAKSATIYNPVIYVFMNRQFRNCILQLFGKKVDDGSELSSA
SKTEVSSVSSVSPA*
The protein is manufactured from 1234 exon bases/letters, which results in a protein of 364 amino acids after being "printed" by the ribosome.
The 1234 exon bases is obtained after 14783 bases were copied to pre-mRNA by the RNA Polymerase copy machine.
The Spliceosome then removed 5 introns with 13549 bases in total.
That means only 1234/13549 => 9.1 % of the original bases are being used for the protein coding.
Here is a 2D representation of the "Red sensitive" Opsin gene in the genome on chromosome X: (The exons are highlighted with red rectangles around them)
long-wave-sensitive (OPN1LW), mRNA.;
uc004fjz.4
(Full gene position = chrX:153409725-153424507)
Exon 1/6 length:145
Intron 1/5 length:6258
Exon 2/6 length:297
Intron 2/5 length:1988
Exon 3/6 length:169
Intron 3/5 length:1467
Exon 4/6 length:166
Intron 4/5 length:1554
Exon 5/6 length:240
Intron 5/5 length:2282
Exon 6/6 length:217
Transcript Length: 14783
Exon bases length: 1234
Intron bases length: 13549
If one could magnify the DNA bases and look at what it would take to recognize and remove the introns and the splice together the exons, you would realize that the processing performed by the Spliceosome does not differ much from text manipulation in a computer program.
In the following pictures I show 2D representation of the exons of the OPN1LW gene coding for the Red Sensitive Opsin.
The regions marked in BLUE represent the bases which are part of the exons.
Intronic bases which are not part of the exons are indicated with:
Yellow = G (Guanine)
Orange = C (Cytosine)
Dark blue = A (Adenine)
Black = T (Thymine)
The codon where the protein translation would start in the Ribosome
is shown in green as ATG
The stop codon which codes for a stop signal is shown in red as TGA.
Splice start and end recognition sequences are marked with a rounded red rectangle around them.
Since this protein is encoded on the positive DNA strand, bases are read in the direction of increasing base position.
Let us first look at the first exon of the gene. This is the first part of the what will be transcribed by the transcription machinery. The promoter sequence normally lies about 35 bases upstream before the start of transcription. Although many genes do not conform to this rule, you can see the TATA box sequence at position -37 = TATAAA
(You can locate it by counting 37 bases backwards from the start of the blue area in the gene)
This is where the TBP Transcription initiator protein binds to "mark" the start of transcription. The TBP protein forms part of what is know as the Transcription initiation complex. The DNA is also bent by this complex to facilitate the binding of the RNA Polymerase.
This sequence is like the "header" for the gene. The RNA Polymerase will transcribe the entire gene until it gets a termination signal (via the synthesized RNA secondary structure "brake"). The transcript then gets ejected from the Polymerase and will then be edited by the Spliceosome.
Also notice that the ATG translation start codon falls in the first exon (which is the blue regions)
Exon 1 length:145
Intron 1 length:6258 (This is represented by the bases which are NOT marked in light blue)
When you look above in the sequence just after the first exon (part of the gene), one sees the recognition sequence for the start of the part to be removed i.e. the intron: GTGAG
This tells the Spliceosome where to "cut" the film during the editing process. The cut is made just before the GT. (In reality it would be before the GU because in RNA Uracil is used in stead of Thymine, but I prefer working with DNA letters).
Something else to notice is that there is no AG before the start of the first exon (as AG always marks the end of an intron, NOT the start of a gene). There are no introns before the first exon. Introns only exist inside the transcript of a gene after the end of the first exon.
Below you can see the last part of the first intron which is 6258 bases long in total (not all of the bases are shown), and as you can see its end is clearly marked by the letters AG just before the start of the second exon. This embedded GT....AG code clearly demarcates the introns to be removed and works like "film editing" synchronization points recognized by the snurps (RNA linked proteins). Also notice the highlighted branch point A. It is called a branch point because after the the 5' GU is cut from the first exon, it forms a loop and gets joined in a 3 way junction at the A branch point. Another snurp recognizes the final AG which is cut just after the G. In a similar way the 3' end of the previous exon is then covalently joined to the 5' end of the next exon.
Exon 2 length:297
Intron 2 length:1988
Look at the base sequence GTAAG which marks the start of the second intron above. In this case it is slightly different from the start-of-intron-marker GTGAG used for the previous intron. The Spliceosome has some leeway or freedom in the exact sequence it recognizes and will allow for slight differences. Natural mutations in the DNA could change the base sequence used to mark the introns which would result in an entire intron not being removed from the transcript if the sequence matching was too strict. This would result in a broken protein due to extra non-protein-coding letters being processed by the translation machinery. That is why the Spliceosome (which may match slightly different sequences in different organisms) matches based on a consensus sequence where the total match is considered and not just individual bases.
In the following picture the size of the letters is an indication of how often a specific base can be found at each of the marker positions of the introns. The larger letters means it is quite often found to mark the start and end of introns while smaller letters means it is found less often.
Exon 3 length:169
Intron 3 length:1467
Exon 4 length:166
Intron 4 length:1554
Exon 5 length:240
Intron 5 length:2282
Exon 6 length:217
Above you can see the last exon of the gene to be spliced to the mRNA eventually being exported from the nucleus. The last exon also contains the translation stop signal TGA which results in the asddition of amino acids to the protein chain to stop and the final peptide sequence to be ejected from the Ribosome.
Something else to notice...there is also NO start of intron sequence GTAAG after the final exon, because there is no more introns after the final exon and this would violate the coding scheme.
In many respects the layered coding scheme used in DNA is very similar to the Network Communication OSI model for digital communications, where, in decoding the information, each successive layer removes the header (containing that layer's context info) from the previous layer's data.
In the following picture you can see a diagram of how the Internet Protocol (TCP/IP) works.
In a future blog post I will be discussing the analogy in more detail, but for now I just wanted to give a short introduction.
There are 7 layers of functionality:
Reliable transmission of information accomplished via a hierarchy of functional building blocks each performing a specific part of the communication task by
- Using the functions of lower layers
- Providing functions to higher layers
The Data Link Layer / Ethernet Layer is in the equipment implementing the modems and network cards.
The IP-Layer is normally implemented in software for the different operating systems such as Windows, Unix, Linux, etc.
The TCP/IP layer is one layer up in software which assembles the IP packets into TCP Segments which are strung together to form the final data stream.
The layers are as follows:
7. Application Layer
6. Presentation Layer
5. Session Layer
4. Transport Layer
3. Network Layer
2. Data link Layer
1. Physical Layer
An easy way to remember the layers is via the following mnemonic phrase (in reverse order):
Please Do Not Take Salami Pizza Away
This is how I think it relates to the information in DNA:
| Physical Layer | The DNA genome housed in your 46 chromosomes - secure storage, double redundancy back-up, replication/mirroring of data |
| Data link Layer | Mitotic Cell Cycle control via a Cyclin Dependent process which controls when the cells duplicate and when they divide - Data frame synchronization (When to send/receive frames) 3 ringed paired bases G-C or T-A between the rungs of the DNA ladder, as well as 3 hydrogen bonds in G=C or 2 hydrogen bonds in T-A base pairings - corresponds to using Parity bits (odd/even parity, CRC check) in data communications (This is used by the DNA Damage detection pathway which moves along the DNA to find BULKY parts which it then repairs by cutting it out and replacing it with the correct bases) The double stranded nature of DNA allowing the one strand to be used as a mirror for regenerating the data when one strand got damaged or incorrectly copied-This is done by distinguishing the strand which is new from the one which is old via methylation marks on the DNA. Double redundancy of information in the genome as 2 homologous chromosomes obtained from father and mother act as a template for homologous repair pathway during formation of Holliday junction - correspond to error detection and correction codes implemented as part of the modem to allow for reliable transmission over noisy physical links |
| Network Layer | RNA Transcription Initiation by the Transcription factors - by recognizing the promoter sequence (such as the TATA sequence) of a gene to tell the copy machinery where to start copying the DNA (Stripping off the Data Link Layer header = Promoter and Activator sequences) Transcription termination - by the recognition of termination sequences derived from the RNA self-base-pairing secondary structure. Gene regulation - Transcription factors react to hormones such as growth hormones or Oestrogen to switch certain genes on and off at specific times. Transcription from DNA into pre-mRNA by RNA Polymerase II - reassembly of the mRNA "Packets" from the bases in the "communication channel", i.e. the genes on the chromosomes. |
| Transport Layer | RNA Splicing and editing Spliceosome - Reassembly of smaller exons ("network packets") in the right order into mRNA (which correspond to TCP packets) (Recognizing the intronic headers and footers and stripping out the introns and their headers from the "Network layer" before splicing together the exons) Nuclease enzymes degrading incorrectly spliced mRNA - Recovery from transmission errors by retrying Nuclear pores - not allowing incorrectly spliced mRNA to pass through due to lariat loops preventing them from going through the pores. mRNA editing such as adding a 5' cap and poly-A-tail to the mRNA - this "header" added to the mRNA protects it from being degraded or seen as RNA viruses and the Poly-A-tail determines the lifetime of the mRNA before it is degraded outside the nucleus. (This corresponds to the time-to-live TTL counter in IP-packets which prevents them from circulating the internet forever by decrementing this counter in the packet along each "hop" from source to destination. |
| Session Layer | Provides for protocols for authentication of information such as Nuclear pores - Direction of flow of information by only allowing mRNA to pass through while stopping other molecules. DNA damage detection pathway - Authentication of source of the information (login, digital certificates), Data security and integrity Transcription regulation and RNA interference regulation - When transmission/movement of information will stop/start DNA Mismatch repair pathway, Base and Nucleotide Excision repair, Apoptosis (programmed cell death) - Data integrity (ensures that information (although reliable) was not changed without being detected) |
| Presentation Layer | Ribosome translation (rRNA), Aminoacyl-tRNA-synthetase enzymes, tRNA adapter molecules - This layer translates information between the data format transmitted over the chromosomes (i.e. DNA/RNA) and the data format of the application layer (i.e. amino acids in proteins) It processes the incoming data stream in groups of 3 as codons (code-words) in the Ribosome A,E,P sites, recognizes the start (ATG) and stop (TGA, TAA, TAG) code words and then translates it from one coding domain (DNA with 4 characters) into another coding domain (amino acids with 20 characters) using the Genetic Code as a lookup table. This is similar to the base 2 2 bits in the transmitted data on the internet being converted into ASCII (128 characters) or UTF8 (256 characters). |
| Application Layer | Proteins - the implementation of functions such as enzymes,hormones, transporter proteins,antibodies,structural building blocks,motor proteins,receptors,signalling proteins and storage proteins. In addition to having "headers" in DNA, there are also "headers" in protein in the form of signalling seguences or nor nuclear localization sequences (NLS) which is a specific sequence of amino acids which are sometimes stripped off during protein processing and folding and which is used for protein targeting and acts as an address to tell the cell whether a specific protein should be exported from the cell or moved back into the nucleus where it functions as a protein machine. Even viruses are designed to make use of these signals/ The first NLS to be discovered was the sequence PKKKRKV in the SV40 (Symian/Ape Virus 40) Large T-antigen which directs the virus' protein to the nucleus. |
As you can see, the information in DNA was not simply willy-nilly thrown together, and there exists clearly defined headers containing information for processing at each layer of the communication system. This system has been used to transfer the body recipe information from the first humans across aeons of time up to where we are today. Errors do occur, but there elaborate error detection and correction schemes in place to ensure that the information is accurate.
I will be concluding by including the primary transcript of the OPN1LW gene on the X Chromosome.
Exons are indicated in capital letters and highlighted in yellow, while the removed introns are shown as lower case letters.
Also notice the intron headers and footers used by the Spliceosome machines to remove and edit the pre-mRNA into mature mRNA messages: gtgag.......cag
That follows is the GENE which allows you to perceive RED colours...
CGGCTGCCGTCGGGGACAGGGCTTTCCATAGCCATGGCCCAGCAGTGGAG
CCTCCAAAGGCTCGCAGGCCGCCATCCGCAGGACAGCTATGAGGACAGCA
CCCAGTCCAGCATCTTCACCTACACCAACAGCAACTCCACCAGAGgtgag
ccagcaggcccgtggaggctgggtggctgcactgggggccaccggccacc
cacctgccccgcccaagggaatctctcttctgcacgtccccaccagcaga
gaaggctttctcccatagcttttctgatgacatgaattggggggtcctct
ccaaatctagaaggacaccataatatcgaatatgcattctcaagccacac
aggcttcccagcccctttgagaatccgaggccggggaagagtttatgtgc
tctttctttgtggcccgtagatgagtgtgttcactgctagcgaatgacct
ctcattccacggagtccctcagcttcctggggaagagctgggtctgtctt
tacatttgaagccgaaagaggcaacatactgacacacccaagggaggcgg
gagggtggggaagacagcagcagagggcaagaaacttctagaacttcagg
gtcggcaaagcctgtagcagtcattttgtcaaactccatgatggggccac
ttggcttttggctgcacacctctgggggaagaggctgcattggcgcccag
ggccatctttccattcggagccgtcctgggagagagggctcaggcccaac
agaaagctgaaagctctcatcagggcagcccgagtcctgccattgggagt
tgcccaatccgaaagttttgcacgcaggccctcaaagaagctgaggacac
cagtgaccgccccactcctggccctctccccaggtccctcctccaaacca
aattcctttggtgccttcaagaacatcgtgcaggccgggcacagtggctc
acgcctgtaatcccagcactttgggaggcagaggcgggcagatgacgagg
tcaacagatagagatcatcctggccaacatactgaaacctcatgtctact
aaaaatgcaaaaattagctgggcttggtggcgcatgcctgtagtcccagc
tactcaggaggctgaggcaggagaatcgcttgaacccgggaggtggaggt
tgcagtgagccgagatcacgccactatgctccagcctggccacagagtga
gactcttgtctcaaaacaaaacaaaacaaaaaacaacaacatcgtgcagg
ctgtggtttccagaagccacgccagctccttgattgccaataaacatccc
gctgtggggtggccaggaccgagtgccaattagtgacagagtgcccagac
caaaccggatgaggatcttgcagttgacctcaacatgactgtgcccagaa
tttccttggtggcaatgtcaacagtctcttcctagatgcccccagacttc
atcaatgcatgatgcttcagtgcactcttttcaaatgtcggggtgggttt
tttttttttccacaaaacttcaagcatctactaaagtagagggaggagtg
taatgaactccggtacccatcactcagcttccacggtttcatctcatttc
atctgtgacccctccactaccctttcttcctgattcttggaagcaaatcc
aagacatcacacccttccctctgtaaatctttactatgttcctctaggag
aaaagggctcttctcaatacataaccacaagtcatcatcacaccgacaag
tgtaacagtatttcctgaatagcttcaaatatcctagtagtgttcaaaaa
atgtcatacgtattttcagtctgcttgaatcagggctcaaataaggtcca
cacattcagattgactgatatgccttttgactacctttgaatctagaggt
tccctttctatctccctgcaatttatttgtggaagcaagcaagtcgttca
tgacgtagcctaacaggcccctctgacgttgttcattatgatttttctgt
aaattggtagttgatctgaggatctggccagaggtaggttggatttgttg
gtgtgttttggcaaggagagtgtctcttttctggggtgttggcagctact
gaaactcaatgcccagaccaattaaaccactggggatggaaaatgacggc
attcggacaccttaccctgccttcacctattggtgaccaaaaccttaaca
tcttcacaggtcttcttaccctgagggtatatgccactaggttgtgtagt
aaaccggtgtgtttccagtcccttagaatagtccctctctaagtgatatg
ccactcagtggatatgcatttagcttcatttcttttgttgctgattttca
gagattgctctgtaaatttaaacttttattttactttattttattttttc
gagacagtcttactctgtcgcccaggctggagtgcagtggcgcgatctca
gctcactgcaacttccgcctcctcggttcgagcgattctcctgcctcagc
ctcccgaggagctgggactacaggtgcccgccaccacgcccagctaattt
tttttatttttagtagagacagggtttcaccatgttggccaggctggtct
ccaactgctgacctcaagtcgtccgcccacctcggcttcccaaagtgctg
ggattacaggtgtgagccaccgcccccggccacttaaattttgttttata
attatgtaataaaacagttaaaagtctcaaattaaaatctagaaaagaag
gtgtatttgaagaagtctggcttctctgcgccaccaccgaccgccccttc
cctacctgcctgtatttcctcgaatcactttgcctgggagttgactttga
ttctcttgctcattgcttcatgaaattcagttccagaactttcaggaggg
aggggtaggccatgacaccagctctagttacactggtggcagctcctgtc
ccctcccccactgctgctgggacctgttctctcctttgcccccttgtccc
tgcactgcccaatttggaccgcaagggttgccagggaagggcactggctg
ccttgttttcagaggtcgtagcacctagattgctccagccccttgcactt
gcctgcaggccagagtgtcccaaaccctcccagtctcagctgctcttccc
cagttcacccaaggtacttcccagggaagagctgccgacagtttgggggt
tctctgttcttaggtccatcagcaaccccattgctcccctctgcttcctt
ctgcacggagactgacgccatgcaggtcttcaattgtcaatggtctgtcc
ctgctgctcatactgggggttcctggggagccagtgccaggtatcgggat
tgcagacattgtctgtgggtttccagaagctccttgtgttaggaacatat
ggggcccgtgcacagagggcagcagaggccttgtgggatccagctgtgct
aggggtgagatttatctgtctctcctggccatagccaggaaatccccatt
tttcttaagctagcttgagttgggcttttctaacacacagctaaagaatc
tcttgataaaccttgggactctccatgaggccttatatggcagcaggtct
gtggcttgcaatcccttcaagtaatctgccaaaaacaatgttatgacgaa
ggtccttccaacacaaaaggtgtagagccctagcaaactcctacagaaga
aaaaggagaaataattcgtttgtagtcccagctacttgggaggccaaggt
gggaggatcacttgaagtcaggagttcgagaccagcctaggcaacatagc
cagaccccatctctacagaaataaaaaaaattgccattgtggtaatgcac
ggcttgtagtcccagatactcgagaggctgaggcaggaggatcgcttgag
cccaggaggatcgcttgagcccaggagttccacgttgcagtgagctatga
ttgtgccactgtactccagcctgggtgccagagccaggctctatctctat
ttggtgttgttgttgttgctgttgttgtttttgagacggagtcttgttct
gtcaccccggctggagtgcagtggcgtgatctcagctcaccgcaacctct
gcctcctgggttcaagtgattctcccgcctcagcctcctgagtagctggg
actacaggcgcccaccaccatacctagctaatttttttttcttttgtatt
tttagtagagacggggtttcaccatggccaggctggttttgaactcctga
cctcaagtgatccaccccctcggcctcccaaagtgctgggattacaggcg
tgagccaccgctcccagccgactgtatctctaaatatataataatcataa
tcataatcaggacagccgtcatattggattagggcccaccctaatgacct
catttaaacttggtcatttctgtaaagaccctatctccgaaaacggtcac
attctgaggtattggggttaggactccaacatatgaatttgggtggggac
acaattcaactcataacacattcattgattaatttcctcattcatttatt
ttctgagcatctattgtgtgctggacactctgtgaggatgaatgagtcaa
agtctctggtgagaaagacaagacttgtacttgtgtctcagtactggctg
ctataagaaattaccaggctgtggccgggcgcagtggctcacgcctgtaa
tcccagcactttgggaggccgaggcaggcggatcacgaggtcaggagatt
gagatcatcctagctaacatggtgaagccccgtctctactaaaaatagaa
aaaattagccaggcgtggtggcgggcgcctgtagtcccagctactcagga
ggctgaagcgggagaatggcgtgaacctgggaggcggagcttgcagtgag
ccgagatcgtgccactgcactccagcctgggtgacagagcaagactccgt
ctcaaaaaaaaagaaaaaaaaaagaaaagaaaaataaattaccaggctgt
gcatggtggctcatgcctgcaatcccagtactttgggaggacaaggcagg
aggatctcttgaggtcaggagtttgagaccagcctggtcaacatggtgaa
accccatgtctactaaaagcacaaaaattagctgggtgtggtggtgggtg
cctgtaaatcccagctactactccggagctgaggcaggagaattgcttga
acccaggaggcggaggttgcagtgacccgagatcacatcactgcactcca
gcctgggtgacagagcaagactttgtcccaaaaaaaaaaaaaagaaaaga
aagaaaggaaaagaataaaagagaaattaccatagattgggtggctttta
aatgataaatttatttctcacagctctggaggctggaagtcagggtgcta
gcgtggtgggctctggcgaggaccctcttcctgactgcagattgccaaca
actcattgtatcctcacatggaagaaagagagctagagagcactctaggg
actctttttcttgtttgttttaattaaaaaaaaattttttttacatgggc
atgccatgttgcccaggttggatttgaactcctgggctcaagcaaccctc
cagcctcagcctcccaaagtgctgggattacaggcatgagccaccattcc
cagctaatttgggctgttcccaaaggctcaagtgatcctcccacgttggc
ctcctgagtagctggggctacaggcgtgagccaccatgcccagcttctag
gacctcttttataagggcactaatcccattcatgagggccccactcactc
tgcacacatgacctaaatgacctgccaaaggccccacctcctaataccat
caccttgggggttgggatttcaacacagaaatttatggggggcacgtaca
ttcagatcatcatgaacagtaactcctatgtgtgacagaaggtgacagag
gtgggtagtggtcttcccctcaagggggtgagttgccactagctggggaa
tcttctggaaggcaaatgcatatgagctgggctttacaggaggcaagcgt
ttctctatggaagggcagaggactgtgggaggtaggaggtggggctgggg
caaagggaagaggggagcagggaagtggggtgactgcacactgggagtgg
ggaatcagatggaggagacgatgaggagttctgttaagttcaagatgcca
gtgccagtgaccagcgggcgatggtctctggcttgagggacaggatggag
gggagactgtctgaggatggacaaagctggagggaaacagccaattgcaa
aggcaggagggcggaagggggaggggagaggtgggatcagcactggtata
gacaggcggtgctgcagcccagctcctctctctcctctgcctcctgccct
cagGCCCCTTCGAAGGCCCGAATTACCACATCGCTCCCAGATGGGTGTAC
CACCTCACCAGTGTCTGGATGATCTTTGTGGTCACTGCATCCGTCTTCAC
AAATGGGCTTGTGCTGGCGGCCACCATGAAGTTCAAGAAGCTGCGCCACC
CGCTGAACTGGATCCTGGTGAACCTGGCGGTCGCTGACCTAGCAGAGACC
GTCATCGCCAGCACTATCAGCATTGTGAACCAGGTCTCTGGCTACTTCGT
GCTGGGCCACCCTATGTGTGTCCTGGAGGGCTACACCGTCTCCCTGTGTG
gtaagccagtcggggcccaggctcggcggaaaccactcattcaccctgca
agctcctccagccacctcatgatgatcggggcccagctgctcctgtaggc
ctgtctccctccacatctgcgcctcacatccatatactgaagggttctgg
aggcttccatctgaacactcacattaaattcagctcccttgagtcaaaca
taccctgagttcctactcttgagtcaggctctgcccggggacagccagtt
tggagctgtggggctggtgtgggaggagacagatacagagctagacaacc
ccagaacagtaggggggcggggactctgggcaccctggacagaactcccc
tgcaattagggatgcctgctctttcagctcgccagcatctgcttttcccg
gaggagacacaattcccagatcctctccccatccccatcactaatatctc
tgtgggccactattccgctcaggtcaggagacagtggccgagaggtacta
gcgtgccaggctctgtgctaaggagggggccctatagccagacggcaacc
acacagtaccatcatcagtcctctcagacaagaaggggcctggggcaggt
ggtggaggagcggctgggagcagtttgtggttcgagtggatagagtacca
ccaagcagccgtggctgctggacacgaggtgggcaggcccaggtctcaga
ggcctcagacgtcatgcccaggagctgggactttctttcaggaggaggag
accccacatccagcagcagcagctcctgctcttgcctccccaccactctt
agcagcctccccaaccccaccccgttaactgcctcaaattgtacccacga
tggcccagaccagagagggtgcttgtccaagtcccggcactaccccgata
gtgtagaaggggagccaagggaaggtcaggcagagaaggtccatccccag
gtccgagtgctctctgcagcaggcatggcctcggtggtcacacgaccctt
cccgagtgccccccctgcatctccgcccacgtctgtctccgtttctgcca
tggtctcccgctcacccttgcctctgctcatggtctgttcttgggtcagt
caggtgccaagcagccagcacttccccaccacttttggtccacggatgcc
cttggccatctgggaagcctgtggaccccatctcaggagaatttttgcaa
acgcataaaatgagacccataggattacaaaggcagcaaattatactgaa
atacagttatcaaagtattaaacattcatcagtaacatagtctttagtta
aaagcatttactggccaggctcatacctgtaatcccagcactttgggagg
ctgaggtgggaggactgcttgcctccaagagtttgagaccagcctgggca
acatagtgagacctcttctctacaacaaataaaaacagctgggcgtggtg
gcacaccagtagtcccagctactcaggaggctcaggcgggaggatcgctt
gagctctggaggtcaaggctgcagtgagctatgatggcaccactgcactc
agcctgggcaacagagtgagattctgtctcaaaaagtaaataaaaataaa
agcatgtgttaaacgtattagtgacaccactcagtattaaggtattaaat
aacaggatcccgcctgacaaccactgttatttcagagtagtgatgaacat
aagtggtattcgaactctctgccacctctatgaattgacaggaaaacatc
tgtgacctctcttgctgaccgagtcacgggtactgctaatactgccacgt
tcataatggaaggaaatgcccagtgtctgttcgaggttggtggaaagaaa
gatgtcgttttttccacctcagtccgtggagccctgaattctgtgtgcag
acgtttggggtctaagcaggacagtgggaagctttgcttcccacctttgc
tttggctcaaagccctcatctgtctgctctccccatagGGATCACAGGTC
TCTGGTCTCTGGCCATCATTTCCTGGGAGAGGTGGCTGGTGGTGTGCAAG
CCCTTTGGCAATGTGAGATTTGATGCCAAGCTGGCCATCGTGGGCATTGC
CTTCTCCTGGATCTGGTCTGCTGTGTGGACAGCCCCGCCCATCTTTGGTT
GGAGCAGgtaagggtgcgaggacgcaagatggagtgggcagggtcagact
ctgtgaccttaaggcaaatcacttcctttctctgggcccctctgagcgtg
caatgtctatcaatgtatgaatgtggctgcaacataggaaaggctctgtg
gtccccgaacctctggaaacatatttatcccaagcacgatcaggtcacag
gcgcacacggagctcaggccatcagcacagctgtcagtgaacgcatagcg
tgtttgcattccaggtctctttcttgcacacgctgccgcaccacgccccc
cacctttcagaggctgcttgggtcatagatccacctgggcctacagagca
catgtcctggccaggccaagcaagtggctcaaatgtttgattggagtgga
ctgggtgggacagcatttcactgttttatcgacaagctcgtgaataagtt
ctcgtggtgtttggagagggaatgttctttcctcgagaacgttccacaat
tctaggaaacaaaccttgtggaagcctgtctctgtctcccgccctcctca
tgccgccatgccccacacagctgcccgttatcaaacatgtgtggtgagct
gaccctggtggaggctctcccgcgggttatctcatttaatcctccaggcc
actaagtgagcagggccctttatttcagtcatggcctagctgacctcaga
taaaagactcagctcttcatgggtgttctcagaaggtcagggcaagaagg
aacctcacaatccctttgtaaagaaggggagtgattgggaagatgaaaat
gtcctggaagcagatagtggagatggttgcacagcattgtgaatgtacca
aaggtcacaatggtacttttttctttttttgagacagggtctcactctgt
cactcaggctggcacagtgcagtggtgtaattatggctcactgcagcctc
cacctcctgggctcaagtgatcctcctacctcagcctcctgaggagctgg
gcctacaggtgcaccacttcacccagctaattttttttattttttgtaga
gacaagatctcactatgtgatccaggctagtcttgaactcctgggctcga
gcaatcctcctacctctgcctccaaatgtgctgggactataggcgtgagc
cattgtgcctggcctataatggtacattttatgtgatgtgtattttacca
caattcaaaaagaagaaaggcatgacatctaaaaatggacaaggattaac
caaaatcctacccaacggttttgttttgggttgatgaaaatgttctggaa
gcagaggtggtgactgccacagaattgatcacttcaaattgggtaatctc
atgcaacatgaatttcacctcaatttaaaaaaacaaaccccacccgagtt
agcaccgtgcctgggccgggggtcctgggtcaccccaccctgcatcagga
ctggctgccggcccttctctccagGTACTGGCCCCACGGCCTGAAGACTT
CATGCGGCCCAGACGTGTTCAGCGGCAGCTCGTACCCCGGGGTGCAGTCT
TACATGATTGTCCTCATGGTCACCTGCTGCATCATCCCACTCGCTATCAT
CATGCTCTGCTACCTCCAAGTGTGGCTGGCCATCCGAGCGgtaagccccc
cgattcctcctggcctcacccgcctcctgcccctaagctgctctgccctc
aaatgagtccactgagactcctaaactatttttccaaaaatccttagaga
agaggattttacccctataagaaaatattaagatccagcgatgagaatca
ggtgattcctttgggactgtaccagtggctgcaggttcagccccagcccc
gttgtcctcagctctgtgagacgggaaagcactgccactccctccctgga
ggagtccactaagggaacagaggtgtgccttgccccgaccctggacagtt
ctccccggggtggaaaggctgcctttcccacagagtagagtggagcagcc
acatcagcaaatgacacctgcaaatcaaggcgtgtttttatgaggctgcc
accggagtacccttgtccttttcataggctgtggggccgaccaaggagtg
gacccgagagtgccatttgcccccctgacccactctccacctccatgtct
ggccctctgccctgggaagctgatcctgtccacagccgtcaccccccacc
cctagactaggctaccactgggagcccttcaggaagtcagagcaagggag
gagagccaggctggttcttttctgttagcagtgggagccctttcagggtg
ctggctttcctatatgaagctgcctgtgcccacaattggatgggcatgcc
tgccaagctctctctagaggagtctgtgagcctgtgaaaggccccctcac
cccgtcaccttggggtgaaggctcccacaggtacccaaccatggcttcgg
ctgtattagtctgggatggtagagccccagctccacaatgtggccccagc
tctgctgtctcagccatccctgcattccagccctcacactccctctctca
tccccactcatctgcctgccgccagtccctcatccctggcaggtggtggc
tggcctctggcctcccccacagtgcctctgcctggaggccattcgtctcc
ttcctcccagcaggcatgaaggagccaccccaccaaagctgccctcagct
gcctcaccgtgagtccagggcaggatttagtccacagagtggccaacctg
gcctaggaagcctgagggaagtgtatgcattgctctgacactcccatcgc
gcaccccgccagccactgcttttgcctcccccgccatctccaccttgtta
actccttcattctccacgcccagtcatcaatcaaatcaggcctccatgct
caggcctgagcgcaggacaggacagtctgttaagggatcaggtgaagcaa
aggagcttgttagatccagctctggggtcatcttaggccacacctagctg
catgccacctccaattctagaactcccccagggccagcctgaggcagcca
tgtctgcctggggccggctgtgctccactcagggctggaagatggctgct
gggctcctctcctcctccccacaactccctatgcctgggtcacctgcctc
ttgctgccctccaacccccgactcactatccctgtctcccttagGTGGCA
AAGCAGCAGAAAGAGTCTGAATCCACCCAGAAGGCAGAGAAGGAAGTGAC
GCGCATGGTGGTGGTGATGATCTTTGCGTACTGCGTCTGCTGGGGACCCT
ACACCTTCTTCGCATGCTTTGCTGCTGCCAACCCTGGTTACGCCTTCCAC
CCTTTGATGGCTGCCCTGCCGGCCTACTTTGCCAAAAGTGCCACTATCTA
CAACCCCGTTATCTATGTCTTTATGAACCGGCAGgtaagcaacaccatca
gcagatcccactcaaaataccgtgtgccctagaagggtgcagtgatggcc
ccacctggaatcatgtctctgataagaagcccgcggagcatctgggggac
cctccagggaaatgaccgggaaaggctcagcgtgtgacccagccccagcc
agagctccagctggcccttagcagaaggcttaggtgtgccctctggaatc
ctttatagtctcggcctgagggtggcatttcccaaagcgtctgtgtgccg
tgcgctcttcccttccggtggccctagaactatggctgccgagcttcagg
ggctctcctggcgttcagacgctctaggagttggtgagccctaggtacat
ccaccctaggtgtgcccctcttctgttcagactcgacccttctcaacctt
catctctccattttcaaaccgtaacctctggaatttgtcttcctataaga
acaaaagccggccctccttggctacactgaccaagagttcaagagctttc
acgagtttgtgggttagttcaggggggacgtgctgtggtcctgcccagag
gcagcctccttagctggcatattgggcctcagcagcaagctgctcacaca
cctaaatccccccacctcctgcaggttacaggcttcattaaagcgcagct
gtgatgtgacttgatggtggccagaaaggtgtgcagaggcctcccatttc
accaggcccagtccatcccttccactgggctcttccttgcttctccatct
tagagccactcaatggctccagcccctttggctcagctttgactcacaca
agccaagtctgcagagttcattaagggttcattctctctggtaactttta
aatagtaagtaggaccaggcctgcagtggatttccgggaactcgctgtag
cacactgatgcccagagtgtagttctatccctgacccctgtttcctgact
ttcatgaggatcttttttaggtttctggaatcctaaactatcttgccaag
tactgtctttactggattatttccattctcctttccagaactccccctgg
acagggggagacagatgtctgcacttctggacctcaccaggcctcgaact
ttgcttttaccctttccacataattatcctgtcctgccacattctgagag
aattttctggaacgcagttccatgaagacagcaaattttgctcaggacag
agtctggcacacagtgggtgctcaagcagcagctgctgaatggattcctc
agccctatctcccagctcttcagccgagctgattctgctgtttgtcccgt
ttcttatgttattaatttcaaccattatattttttatttttgagagtttt
gatgatagagggagttagagctagtcaagagtaggcctgaaatatttaga
aaatgcctttggtctgggtcctcaaagcattgtggttacttcagggatga
cacaggacatgatttgagacattcatatggcccagatctctttggggtga
agcagcaaagacagacccctcctggtaccggaagacgcttggctggagag
atgaggtaggggctagattgtcattacctaggcctcaccttgccccagat
ccatggactggaaaaaacatgacaaccacatgccttttcattaatattcc
tccgagccgctcaccagacagtctggggacaggtcaccactgccccttag
ctgtcactgtggatgagtgtcatggggctgccgtcacaaactaccacaaa
ctcagtggcttcaaaccacagaaatggattctctcagggttctggaaatc
ttgagtctgaaatcagggtgttggcaaatggaaaggttccctatggaggc
cgggagggagaagcagctgcagggctgccggcagtctttggcgttccttg
actccaaggtgtgtcaccccagtctctgccttcatcttcacgtggccttc
ttccctctgtctgcgtgtccgtgtccaagcgttccttttcttatcaggac
accagtcattcgattagggcccaccctgctccagtgtgacctcatcttaa
cctgaacacatcttttgggggacccacttcaacccagtgtagtcaccatc
aactgctaagtcagatgacatccccgcgtgtgagggagaaataatccaag
ccttcctccatcccccatgggattcggaatgggtgaagggaaggctcggg
cacgtacattcagcacagtgctccacccttccctgctctgctcaataacg
ctttctgtccttccagTTTCGAAACTGCATCTTGCAGCTTTTCGGGAAGA
AGGTTGACGATGGCTCTGAACTCTCCAGCGCCTCCAAAACGGAGGTCTCA
TCTGTGTCCTCGGTATCGCCTGCATGAGGTCTGCCTCCTACCCATCCCGC
CCACCGGGGCTTTGGCCACCTCTCCTTTCCCCCTCCTTCTCCATCCCTGT
AAAATAAATGTAATTTATCTTTGCCAAAACCAA
Maybe the following picture will now make more sense:

Lastly, here follows a video animation depicting the splicing process on a piece of RNA:
I am truly amazed by the complexity and elegance of this communication/storage system embedded in the organisms on earth.
Hope you also find the information I have shared with you fascinating.

















