I have recently become fascinated with the complexity of the human immune system and how the body is able to ward off infections from pathogens that it has already encountered in the past.
The immune system consists of 2 "arms" of protection:
- the innate immune system which relies on genes encoding for Toll (Great) receptors which are hard coded into the human genome allowing it to detect features of pathogens that are common to a wide range of bacteria like the Peptidoglycan in the membrane of bacteria and double stranded RNA in the cytoplasm (which is a sure sign of RNA viruses replicating). These are all protection encoded into us to protect against a wide range of enemies.
- and then there is the adaptive immune system, which is able to learn and adapt to new threats from the environment by identifying the correct B-Cell or T-Cell that will be able to bind to specific invading pathogens that it has encountered in the past and then to activate and multiply it to enormous numbers.
In order to be able to neutralise and bind to the enormous diversity of possible enemies that might invade our body, our adaptive immune cells generate an enormous amount of different immune cell receptors which can bind to every possible enemy that we might ever encounter in life.
I am not an immune system expert, but I have been able to piece it together like this:
When your white blood cells (including B-Cells and T-Cells) are developing in your bone marrow, they are originally created with what is called the germline/unaltered DNA that are also found in all of your body cells. But, during the development of - and T cells, the receptor protein genes that exist on the surface of these cells go through a process where they self-edit their own DNA by introducing directed double strand breaks and re-joins in order to generate an unfathomable amount of diversity in the receptors, able to bind to every possible antigen that they might every encounter in life. They accomplish this with the use of specialized enzymes which recognize very specific palindromic DNA base sequences. A palindrome is a sequence that is normally the reverse complement of itself:
Like:
CACTGTG
GTGTCAC
They reason that it needs to be palindromic is to allow these proteins to be able to attach and bind to these sequences in both the forwards and the backwards direction.
During development they then go through a process of editing they own DNA in certain parts of the chromosomes by deleting some of the original "germ line" gene segments and sticking them together in a very sloppy fashion in order to introduce even more diversity.
B-Cell receptors (or antibodies when they are free floating) consist of a heavy chain and one of two possible light chains (named kappa and lambda). Together they form a protein that is Y shaped and which consists of 2 identical heavy chains and 2 identical light chains.
Similarly, T-Cell receptors (found on Helper T-Cells or Cytotoxic T-Cells) are similarly made up of Beta (heavy) chains and alpha (light) chains. There is also a Gamms-Delta T-Cell receptor made up of a Delta (heavy) chain and a Gamma (light) chain.
The genes coding for the receptors of your B-Cells and T-Cells are found on the following chromosomes:
For B-Cells (Immunoglobulin receptors)
Heavy Chain genes : Chromosome 14
Kappa Light Chain genes : Chromosome 2
Lambda Light Chain genes : Chromosome 22
For T-Cells
Beta (Heavy) Chain genes : Chromosome 7
Alpha Chain genes : Chromosome 14
T-Gamma-Delta-Cells
Delta (Heavy) Chain genes : Chromosome 14
Gamma Chain genes : Chromosome 7
Furthermore, remember that you have actually got two Chromosome 14's. One inherited from your dad and another from your mom. This means that you have 2 possible collections of genes that can form the heavy chain of an antibody and 2 kappa and 2 lambda gene collections as well. This means a B-Cell's receptors may be produced from either the paternal or maternal genes, whichever is able to successfully form a successful receptor.
The gene segments that are recombined are one of 4 possible types:
Variable (V) segments
Diversity (D) segments
Joining (J) segments
Constant (C) region segments
The germ line gene collection might look as follows:
V1-V2-V3-V4-V5--Vx-D1-D2-D3-D4-D5--Dx-J1-J2-J3-J4--Jx-C1-C2-C3-C4-Cx
There are a number of V, D, J, and C segments which make up each receptor gene type. Successive duplication and mutations have resulted in a collection of adjacent gene segments flanked by specific recombination "signals" in the DNA.
Now think of this gene segment rearrangement as a card game where you randomly pick a "card" from each of the packs of playing cards: V-cards, D-cards, J-cards and C-cards.
The resulting rearranged gene will always contain only 1 of each type (with some exceptions made for the Constant regions, which could contain more than one "card"). Even though the germline contains multiple slightly different copies of the same type (V, D, or J), only one of each is selected and the unused ones are cut out.
Each B-Cell or T-Cell precursor cell will go through this process resulting in many millions of possible combinations:
V2-D1-J3-C1
V1-D2-J4-C2
V55-D5-J3-C4
V21-D1-J3-C1
etcetera...
In this way a multitude of possible arrangements will result in a multitude of possible cell receptor proteins that will be able to "stick" to all shapes of pathogen antigens.
In addition to the multitude of combinations, the process will also insert or remove random DNA letters at each of the joining locations between V and D and between D and J. This will increase the possibilities even further as an inserted or removed base might shift the protein reading frame. Which means the 3 letter codons coding for amino acids will be read in a different frame, for example:
IGHD3-10 ; immunoglobulin heavy diversity
chr14:105904497-105904527)
Normally the 3 letter codons will be translated to amino acids as follows:
GTA TTA CTA TGG TTC GGG GAG TTA TTA TAA
Val-Leu-Leu-Trp-Phe-Gly-Glu-Leu-Leu-STOP
V L L W F G E L L
When the frame is shifted as follows by the addition of an extra base, this results in the codons being translated into totally different amino acids:
AGT ATT ACT ATG GTT CGG GGA GTT ATT ATA A
Ser-Ile-Thr-Met-Val-Arg-Gly-Val-Ile-Ile
S I T M V R G V I I
I have used colour coding to highlight the original codons.
This recombination process is achieved by special protein enzymes in these cells which recognise very specific "codes" in the DNA which tells them where to cut and re-join the DNA segments.
These are called recombination signal sequences (RSS), which can take the following formats:
(The V/D/J GENE SEGMENT)-(palindromic 7 bases)-(12 base spacer)-(AT rich 9 bases)
(The V/D/J GENE SEGMENT)-(palindromic 7 bases)-(23 base spacer)-(AT rich 9 bases)
or
(AT rich 9 bases)-(12 base spacer)-(palindromic 7 bases)-(The D/J GENE SEGMENT)
(AT rich 9 bases)-(23 base spacer)-(palindromic 7 bases)-(The D/J GENE SEGMENT)
Also note that the 7 base palindromic sequence is always directly adjacent to the V/D/J segment while the AT-rich 9 base sequence is furthest away from the segment.
The AT-rich sequence will normally partially base pair with the AT-rich sequence in the next gene segment which is being joined.
It is often easier to explain by example:
Start by looking at the GTGACAC on the right next to the T-Cell Receptor Gamma Variable segment gene. This is a partial palindrome:
GTGACAC
CACTGTG
You always find the 7 base palindrome next to the V, D or J gene segment.
That is followed by a 12 or 23 base spacer sequence. The spacer sequence allows the DNA to bend and properly join the DNA helix at the correct number of turns.
This ensures that a 12 recombination sequence containing the 12 bases will only be able to recombine with a recombination sequence containing 23 bases as that will ensure that the DNA helix is rotated the correct number of degrees for the helix to be properly ligated (joined) back together again.
In the middle where the two AT-rich sequences meet, the GTTTTTAGA from the J segment base pairs with CTAAAGTC from the V segment. This is part of the recombination process.
The following figure shows a summary of the B-Cell and T-Cell recombination sequences as well as the various 7 base palindromic heptamers which are directly adjacent to the gene segments.
Click on the link below the figure for a magnified version of the image.
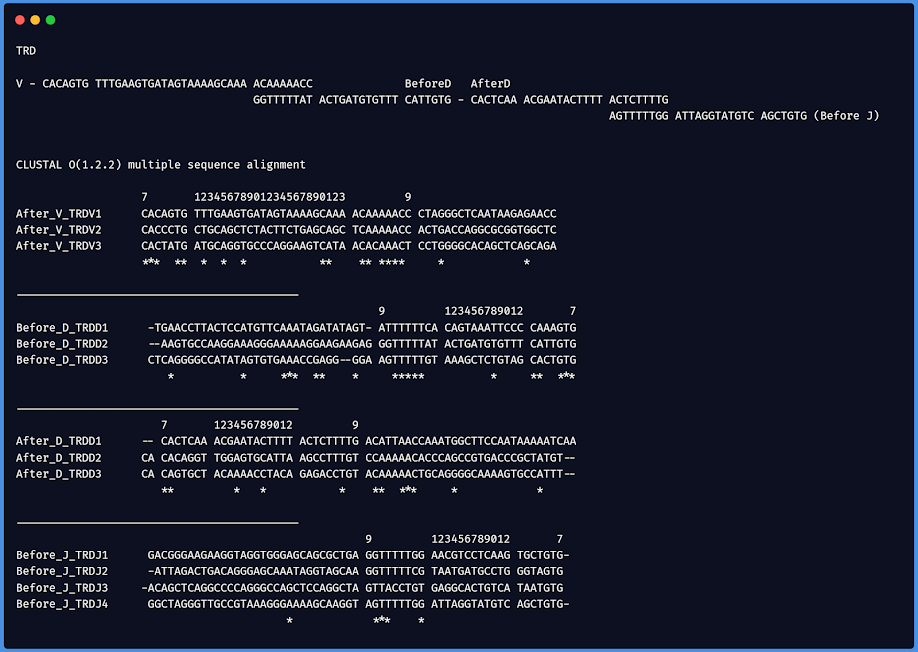
In order to find the "consensus" of the sequences being employed I listed all of the 40-60 bases before and after all of the V,D and J segments of all B-Cell and T-Cell gene segments and then did a multiple sequence alignment on them using CLUSTAL-O. This allowed me to determine how the recombination sequences looked like that is used to recognise where to cut the gene segments and re-join one V, D and J to make the final rearranged receptor coding sequence.
The bases are shown in the order and polarity that they are transcribed and not as they appear on the chromosomes because in many instances like for the B-Cell receptor genes they are actually encoded on the negative strand. This means I will show it in the same polarity as how the gene is translated.
Lets start with the B-Cell receptor / Immunoglobulin Heavy Chain
The following image depicts the Recombination Signal Sequences (RSS) following all of the possible Variable (V) segments of the IGHV genes that the cutting enzyme can choose from. The V segment RSS contains a 2-turn/23 base spacer, which means it has to be paired with a Diversity(D) segment which contains a 1-turn/12 base spacer. A Variable (V) segment therefore has to join with a Diversity (D) segment and cannot directly join with a Joining (J) segment which also contains a 2-turn/23-base spacer.
23-base joins to 12-base (resulting in 3 turns)
But 23-base does not join with 23-base (resulting in 4 helical turns)
Also 12-base does not join with 12-base (resulting in 2 helical turns)
IGHV segments
Above you can see the RSS that goes before the D-segment (the one that recombines with the V-segment), as well as the RSS that follows the D-segment (the one that recombines with the J-segment).
Notice how the 7 base palindromic sequence (CACGGTG or CACAGTG always sits directly adjacent to the D-segment).
IGHJ (the Joining or J segment)
And finally, when we put it all together:
(Notice the overlap at the A or T rich 9 base sequences.)
The Joining segment is then followed by a constant region which can be any one of the following:
IGHM (constant - mu) - resulting in M-Antibodies
IGHD (delta) - resulting in D-antibodies embedded in the B-Cell as B-Cell Receptors
When B-Cells first get activated by contact with the exact antigen which binds to this specific VDJ arrangement, only the IGHM and IGHD constant region is included because there is a stop signal after IGHD. The remaining constant regions are not initially used. This is why IgM and IgD antibodies are the first to be produced since the B-Cell can switch between including the IGHM and IGHD gene regions via alternative splicing. This change is done to the messenger RNA after transcription.
IgM antibodies are usually also the first on the scene to fight pathogens.
When IGHD is included during alternative splicing, a hydrophobic region ensures the antibody stays in the B-Cell membrane. These antigens are then pulled into the B-Cell (for digestion into shorter peptides inside the phagolysosome). After digestion these short peptides are then loaded onto an MHC2 protein complex on the surface of the B-Cell and presented by the B-Cell to T-Cell Receptors on Helper T-Cells.
Lets go back to the order of B-Cell and T-Cell activation.
B-Cells with the correctly combined VDJ receptors can bind to the recognised pathogens that are covered in complement proteins and get partially activated to start producing IgM antibodies,
Simultaneously, dendritic innate immune cells will patrol the areas of infection and take samples of the foreign proteins by chopping them up and loading them onto their MHC2 surface proteins. These dendritic cells then migrate to the lymph nodes where they will present these antigen peptides to the T-Cells. When they meet up with a T-Cell which has also gone through the same process of VDJ recombination, and this recombination just happen to be the "lucky" rearrangement that can strongly bind to the peptides presented in the MHC2 proteins on the surface of the dendritic cells, the T-Helper Cell will get activated if some more conditions are met. (CD4 Proteins on T-Helper Cells is needed to check if the peptides are actually presented in your own MHC2 proteins before the T-Cell can be activated.)
Simultaneously, the dendritic cell (which was originally alerted to danger via Toll like receptors) will give a second danger signal to the CD28 receptor on the T-Cell to tell the T-Cell that the antigen being presented is actually from a pathogen and not your own body proteins.
When the peptide binds to the rearranged T-Cell receptor only in the context of an MHC2 protein on the surface AND the second danger signal triggers the CD28 receptor, only then will it be fully activated and will start multiplying to ensure that this fortunate combination of VDJ genes will proliferate to then go and fully activate the B-Cells recognising this same pathogen (albeit different parts of it).
If a partially activated B-Cell does not meet a T-Helper Cell that can recognise the same antigen, it will eventually die after a certain number of hours. But, if the B-Cell is successful in meeting up with a T-Cell recognising the same antigen (due to a very fortunate VDJ rearrangement), the T-Helper-Cell will in turn secrete cytokines that will tell the B-Cell what type of B-Cell to turn into depending on whether it needs to fight viruses, bacteria, cancer cells or even parasitic worms. This means the B-Cell will again start to swap out constant DNA segments to turn its antibodies' tail section (called the Fc section) into the correct type which is able to dock with innate immune cells like Neutrophils or Mast cells that have Fc-receptors on their surface.
T-helper cells will also stimulate the B-Cell receptor to introduce more mutations to try and fine tune the antibody via a process called affinity maturation: which is basically a fancy word meaning the best fitting antibodies will be stimulated more to continue living while antibodies that are less able to collect and inject antigens for presentation on their MHC2 will not be stimulated and die out. This again happens via recombination of the constant regions of the B-Cell.
Possible remaining constant regions are: (in order of occurrence after IGHM and IGHD)
IGHG3 (gamma 3) - producing IgG3 antibodies
IGHG1 (gamma 1) - producing IgG1 antibodies
IGHEP1 (epsilon p1) - producing IgGP1 antibodies
IGHA1 (alpha 1) - producing IgA antibodies
IGHGP (gamma p) - producing IgGP antibodies
IGHG2 (gamma 2) - producing IgG2 antibodies
IGHG4 (gamma 4) - producing IgG4 antibodies
IGHE (epsilon) - producing IgE antibodies
IGHA2 (alpha 2) - producing IgA antibodies
Each of the antibody classes has their own advantages and disadvantages:
IgA - lines the epithelium and is used to drag pathogens out via the digestive system. They have no Fc portion in order that innate immune cells cannot bind to them and cause inflammation in the gut.
IgE - can be bound at their Fc receptor (constant region side) by Mast cells and Eosinophils to fight parasites like worms
IgG antibodies circulate the blood and binds to viruses and bacteria (and can also pass from mother to baby via breast milk). Cells of the innate immune system like Natural Killer cells and Macrophages can bind the Fc portion to kill enemies via antibody mediated killing where IgG antibodies are used to specifically "mark" (opsonise) enemies to macrophages, neutrophils and natural killer cells via antibody mediated killing.
The mechanism of Class switching is dependent on the cytokines being secreted by the T-Helper Cells which then stimulate transcription factors that bind to specific promotor/switch sections of the constant regions. This results in the appropriate segment being transcribed, which means the DNA at those segments will be pulled apart and exist as single stranded DNA, which are then targeted by a DNA cutting enzyme which can cut single stranded DNA. Only the segments being transcribed will be cut and because they are single stranded at this stage, this will result in double stranded breaks in those segments which will then be repaired via homologous DNA break repair. Because there is a lot of similar repetitive sequences in-between these constant regions, this is also where recombination will then happen. (But only to the segments that was activated by the transcription factors binding to the appropriate promotor sequences)
When looking at a DotPlot of the region between the IGHG3 (gamma 3) region one can see the repetitive sequences.
Here is a DotPlot for the region just before the IGHG1 (gamma 1) region, with equivalent sequences colour matched.
When we look at the Kappa and Lambda light chains of the B-Cell Receptor
The light chains of the B-Cell receptors do not have a diversity (D) segment and here the Variable (V) segment is able to directly join with the Joining (J) segment because the J segment has a 12 base (1 turn) RSS that allows it to work.
IGKV
IGKJ
IGLV
IGLJ
The T-Cell Receptors are generated in a similar fashion to the B-Cell Receptors/Antibodies via VDJ recombination.
The difference is that the T-Cell receptors stay attached to the T-Cells and do not detach and go into the rest of the body as antibodies. B-Cells receptors are also able to bind to a greater range of antigens than T-Cell receptors which can only bind to small peptides (of 8-15 amino acids) and only when it is presented in MHC1 and MHC2 proteins on the surface of other cells.
Something that I have not mentioned yet is that T-Helper cells (also called CD4 positive cells) recognise peptides presented in MHC2 proteins and T-Killer/Cytotoxic cells (also called CD8 positive cells) recognise peptides presented in MHC1 proteins. This is because CD4 proteins can bind to and recognise a part of the MHC2 protein while CD8 proteins can bind to a part of the MHC1 protein.
MHC1 proteins are found on almost all body cells except red blood cells and this means that almost all body cells can present antigens from pathogens to Killer/Cytotoxic-T-Cells, which will then respond by instructing those body cells to gracefully commit suicide via programmed cell death (which is basically the body's recycling process). The most important reason why killer T-Cells are needed is because B-Cell antibodies are not able to bind to pathogens inside body cells. Antibodies exist inside bodily fluids like blood and lymph and that is why it is considered part of the humoral immunity. Cellular immunity is provided by Killer T-Cells because they can peer into infected or cancerous cells and eliminate them. When body cells first gets infected by a virus, the interferon that gets secreted will tell cells to all increase the number of MHC1 proteins on their surface to make them more "transparent" to Killer-T-Cells. All cells will then present peptides from all the host proteins as well as virus peptides on the surface of the cells. The Killer-T-Cells will "inspect" the cells for matching antigens and kill the ones which bound to their T-Cell receptors successfully.
MHC2 proteins are only expressed on dedicated antigen presenting cells (APC) such as dendritic cells, macrophages and B-Cells. These APCs will digest antigens into small peptides and then activate Helper T-Cells. Helper-T-Cells will in turn activate B-Cells and Killer-T-Cells.
Killer-T-Cells should not be confused with Natural Killer cells. Natural Killer cells are part of the innate immune system and they indiscriminately will kill all cells who do not have MHC1 proteins on their surface or which have been "marked" for destruction via antibodies binding to parts of viruses leaving the infected cell. Natural killer cells can bind to the Fc portion of those antibodies via Fc receptor on the Natural killer cell's surface. Natural Killer cells are necessary to close a loophole that gets exposed because of the tendency of some pathogens (including SARS-Cov-2) to try and hide from the immune system by causing cells to stop expressing their MHC1 proteins on their cell surfaces. Without these MHC proteins Killer-T-Cells can then not scan the pathogen peptides with their T-Cell receptors. Having Natural Killer Cells to also kill cells which are NOT presenting MHC1 closes this loophole.
The T-Cell Receptor Beta and Delta is analogous to the Heavy chain of B-Cells
T-Cell Receptor Beta (part of Alpha Beta T-Cells)
TRBV
TRBD
TRBJ
T-Cell Receptor Delta (part of Gamma Delta T-Cells)
TRDV
TRDD
TRDJ
The T-Cell Receptor Alpha and Gamma is analogous to the Light chain of B-Cells
T-Cell Receptor Alpha (part of Alpha Beta T-Cells)
TRAV
TRAJ
T-Cell Receptor Gamma (part of Gamma Delta T-Cells)
TRGV
TRGJ
If we summarise the 7 base palindromic heptamers:
(Showing that there is a strong correspondence between the T and B- Cell heptamers)
B-Cell
IGHV CACAGTG
IGHD CACAGTG CACTGTG
IGHJ CAATGTG
IGKV CACAGTG
IGKJ CACTGTG
IGLV CACAGTG
IGLJ GTCACAG
T-Cell
TRBV CACAGTG CACAGCC CACAGCG
TRBD CATTGTG CACAATG
TRBJ GGCTGTG CACTGTG
TRDV CACAGTG CACTATG
TRDD CAAAGTG CATTGTG CACAGGT
TRDJ GGTAGTG AGCTGTG
TRAV CACAGTG CAGACAG
TRAJ CACTGTG
TRGV CACAGTG
TRGJ CACTGTG CAGTGTG
For more on the subject I implore you to go read the book "Immune" by Philipp Dettmer. Or listen to it on Audible.














No comments:
Post a Comment
Please leave me a comment